牧草开花的分子机理研究进展
English
-
开花是植物重要的生长过程,适宜的开花时间可以使植物有效避免恶劣的自然环境,是成功进行生殖生长的重要保证[1]。开花期是植物重要农艺性状,在牧草草产量、种子产量、饲用品质、混播草地利用价值等方面有举足轻重的地位[2]。提前开花会限制牧草的营养生长,延迟开花则会限制种子的发育或使植株在遭遇恶劣天气前已经发生衰老[3]。牧草开花后饲草品质呈快速下降趋势,草牧业中可以基于不同生产需要,培育不同生育期的早、中、晚熟品种。早熟品种更适合种子生产,晚熟品种在混播草地中可以大幅度提高草地生产能力和利用效率,对推动草牧业发展具有深刻意义。
牧草开花时间的调控对于其生存繁殖及利用价值至关重要,受到自身发育规律和外部环境条件的双重影响[4]。随着分子生物学的发展,模式植物拟南芥(Arabidopsis thaliana)、水稻(Oryza sativa)、小麦(Triticum aestivum)、大麦(Hordeum vulgare)等一年生作物的开花分子机理及调控网络已进行了系统深入的研究,取得了重要成果。而牧草基本上是多年生、异花授粉植物,生产利用方式和作物具有明显区别,但其开花分子机理研究较少。本文就植物开花分子机理研究最新成果进行了综述,以期为牧草开花基因挖掘、功能解析和分子机理研究提供参考,加快其分子育种进程,提高育种水平和效率。
1. 开花调控途径
目前,通过拟南芥研究发现了多条开花调控途径[5](光周期、春化作用、自主促进和赤霉素途径等)以及相关200多个预测基因[6]。光周期和春化与环境外源信号有关,通过光照、温度调控植物花期。而自主促进和赤霉素途径与植物本身相关,是根据自身生长、发育和内源性激素水平调节植物花期的[7]。光周期和自主促进途径在双子叶和单子叶植物之间非常保守,但春化途径出现较大差异,其关键在于启动抑制物的性质不同[拟南芥中的FLOWERING LOCUS C(FLC)和谷类中的VERNALIZATION2(VRN2)][8-9]。
1.1 光周期途径
光周期途径是指植物在感受到昼夜变化后,调控自身以促进开花的过程[10]。根据光周期的不同,可以分为长日照和短日照开花植物[11]。多年生黑麦草(Lolium perenne)是短日照开花植物,而长日照开花植物则有拟南芥、小麦等。长日照(LDs)情况中,在光周期作用下拟南芥等提前开花,短日照(SDs)情况下光周期受到抑制,拟南芥开花延迟。但是Fernández等[12]发现在短日照条件下,拟南芥在CONSTANS(CO)相互作用下可以通过升高温度使光周期更加敏感,进而促进开花。
在模式植物拟南芥属中,CO基因在光受体和生物钟的相互作用下特异性调控植物光周期途径,表现出鲜明的动态变化,GIGANTEA(GI)基因同样首先在拟南芥中鉴定,并且是被子植物中光周期开花的重要调节者。研究证明,CO是连接光信号与 FT基因的枢纽。在光信号影响下,CO蛋白稳定性遭到破坏后将光信号传递到成花信号枢纽 FT 等基因处。此外,CO基因表达也是植物开花时间与生物钟之间的纽带,生物钟借助节律基因GI调节CO的表达,促进花分生组织特性基因APETALA1 (AP1)、LEAFY(LFY)和CAULIFLOWER(CAL)的表达,激活FT基因,调控植物开花进程[13]。
禾本科植物GI功能已被证实,特别是短日照条件下开花的草本植物。在水稻中,OsGI通过控制OsCO(OsHd1)的表达来调控开花时间,使其在短日照条件下开花提前,在长日照条件下开花延迟[14]。在玉米(Zea mays)中,ZmGI1基因影响植株的发育并抑制长日照条件下的开花[15]。
在多年生黑麦草中,LpCO(与LpHD1相同)属于锌指蛋白,含有保守CCT结构域,是光周期途径中重要的转录因子[16]。LpCO的调节机制与拟南芥CO基因类似,转录表现出昼夜振荡,在长日照条件下,诱导下游基因FT以促进多年生黑麦草的开花。多年生黑麦草的LpGI基因,与单子叶植物、真核生物的GI基因结构整体保守,与草甸羊茅(Festuca pratensis)的GI蛋白最为类似。Gagic等[17]推断LpGI极有可能与拟南芥GI直系同源,并参与黑麦草光周期开花时间控制。
但是,研究人员还发现,对于温带豆科草本如地三叶(Trifolium subterraneum),其日间长度和光质量响应的整合可能不受CO-like(COL)基因的调控[18]。
1.2 春化作用途径
春化作用指植物经过一段时间持续的低温处理,从营养生长向生殖生长转化并开花的生理现象。小麦、大麦和其他温带牧草在春化反应方面表现出广泛的遗传差异。根据牧草春化特点,可分为冬性、春性以及弱冬性3种类型[19]。其中冬性品种若未春化,植株则维持在营养生长阶段,或营养生长转化为生殖生长过程持续,致开花延迟。春性品种对低温反应迟钝,弱冬性品种只需经历短暂的春化处理。上述差异体现了重要的农艺价值,因为春性和冬性品种的最佳播种日期不同,适应地区亦不同[20]。
表观遗传调控在春化途径中扮演重要角色,主要通过对表观遗传信息即DNA序列以外的遗传信息进行调控,如DNA甲基化、组蛋白修饰、染色质重塑、非编码RNA调控等。在拟南芥研究中,春化过程与Polycomb Group (PcG)介导的组蛋白甲基化有关。PcG是动植物发育过程中的表观调控因子,包括Polycomb Repressive Complex 1 (PRC1)和Polycomb Repressive Complex 2(PRC2)两种复合物。PRC2复合体决定H3K27me3的形成,当H3K27me3积累到一定程度时,染色质表观呈现抑制状态,此时靶基因转录水平会显著降低。因此春化发生后FLC表达水平降低,其主要原因是长时间低温处理后FLC染色质从转录激活状态向转录抑制状态转变,且伴随着H3K4me3、H3乙酰化等转录激活标记的去除和H3K27me3、H3K9me3 等转录抑制标记的积累。可见,春化对FLC等开花抑制因子的转录抑制受到PRC2复合物调控[21]。
FLC基因一般通过抑制FT 和SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1(SOC1)的表达来推延植株开花。因此,FLC 基因表达的下调解除了对FT和SOC1基因表达的抑制作用,进而加速了植物的开花进程。
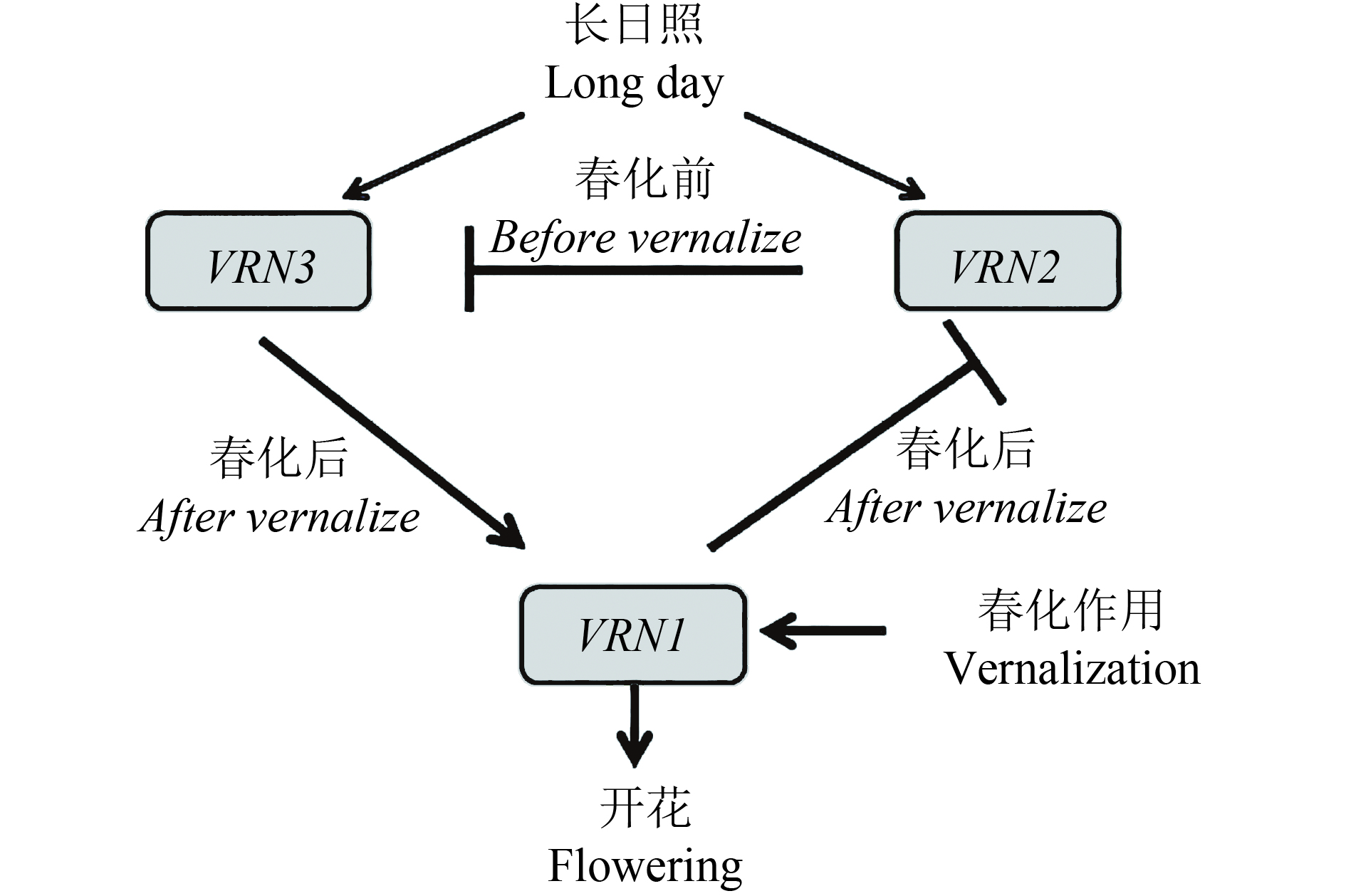
目前,发现麦类作物中春化作用途径控制开花的基因主要有VRN1、VRN2、VRN3以及VRN4,他们通过各自的方式一起调控麦类作物开花[22]。VRN1、VRN2 和VRN3之间相互作用形成一个反馈调节环(图1)[23]。春化前,VRN2 约束VRN1、VRN3的表达。春化进行时,VRN1活性随之上升,VRN2活性随之下降,此时VRN2的表达受到VRN1的抑制。春化作用加剧时,VRN2 对VRN1、VRN3的抑制作用完全消除,VRN1、VRN3的表达加速,到达诱导开花需求的临界点[24]。可见VRN2对开花具有负调控作用,是开花抑制因子,所以麦类作物主要抑制VRN2基因的表达来促进开花[25] 。目前,VRN1、VRN2、VRN3 基因均被克隆,VRN4基因仅存在D基因组(VRN-D4)中,且未被克隆。Kippes等[26]发现VRN-D4属于春化途径一部分,处于VRN1、VRN2和VRN3基因上游,是VRN1、VRN2和VRN3基因的反馈调节环的一部分。
![]() 图 1 冬小麦VRN1、VRN2和VRN3反馈调节环图1显示光周期敏感的冬小麦中VRN1、VRN2和VRN3基因之间的相互作用,箭头表示促进作用,“T”形表示抑制作用。Figure 1. Winter wheat VRN1, VRN2 and VRN3 feedback regulator ringsFigure 1 showing the interactions among VRN1, VRN2, and VRN3 genes in a photoperiod-sensitive winter wheat. Arrows show induction, “T” show repression.
图 1 冬小麦VRN1、VRN2和VRN3反馈调节环图1显示光周期敏感的冬小麦中VRN1、VRN2和VRN3基因之间的相互作用,箭头表示促进作用,“T”形表示抑制作用。Figure 1. Winter wheat VRN1, VRN2 and VRN3 feedback regulator ringsFigure 1 showing the interactions among VRN1, VRN2, and VRN3 genes in a photoperiod-sensitive winter wheat. Arrows show induction, “T” show repression.1.3 自主促进途径
自主促进途径与植物自身有关,是通过植物体内自身特性调控开花的方法,主要通过抑制FLC来促进开花[27]。目前该途径发现的关键调节因子有FCA、CK2、PP2A-B’c、FLD和REF6等基因。依照编码的蛋白可以划分为RNA结合蛋白和染色质重塑因子。其中RNA结合蛋白有FCA等基因,是以mRNA 水平来调节FLC基因的表达的;染色质重塑因子包括FLD和REF6等基因,其编码的蛋白对 FLC 染色质进行修饰。此外,还有CK2和PP2A-B’c介导FLC的翻译进而修饰调控开花[28]。总之,自主促进与春化途径一样,通过控制FLC基因的表达来促进开花,这使得春化作用途径与自主促进途径通过FLC基因连接在一起[29]。最近研究发现,TATA结合蛋白相关因子(TAF)识别基因的核心启动子,是TFIID复合体中的一般转录因子。TAF15b在自主促进途径中通过上调FLC基因影响拟南芥开花时间[30]。
1.4 赤霉素途径
赤霉素途径主要依靠外源施加赤霉素替代春化作用促使植株开花。Yabuta和Hayasi[31]从赤霉菌培养基滤液分离出了一种致使水稻患恶苗病的物质,同时发现该物质对于植物节间生长具有促进作用,因而命名该物质为赤霉素(gibberellin acid, GA)。赤霉素是一种植物激素,合成于植物的芽、嫩叶等。其机理为刺激茎、叶的伸长生长,抑制块茎形成,抑制侧芽休眠,抑制衰老,提高生长素水平等[32]。赤霉素通过激发开花基因 LFY 的转录活性,刺激上调 LFY和 SOC1基因的表达,进而启动开花[33]。在Xue等[34]的研究中,编码赤霉素2-β-双加氧酶1的基因在水稻胚中高度表达,红三叶(Trifolium pratense)的早花和中花阶段,赤霉素2-β-双加氧酶也高度表达。
2. 开花途径关系
开花途径各自孤立,但又彼此联系。研究表明,开花途径在节点基因的调控下关联在一起[35]。其中在拟南芥的开花研究中,发现开花途径与内源、环境信号相互作用,使这些信号向开花整合基因FT、LFY、SOC1、FLC和CO等基因聚合表达,共同调控开花进程[36]。
FT 基因位于不同开花途径的交汇处,在植物开花调控中起关键作用[37]。联合光周期、春化和自主途径的信号,调控植物开花。LFY基因既调节花序向花分生组织转变,又在光周期、春化、自主促进和赤霉素途径共同作用下控制开花时间[38],是整合外部环境和内源信息的重要开花控制因子[39]。SOC1是编码MADS-box的转录因子,在花器官发育相一致的叶片和分生组织中表达[40]。在两个SOC1启动子处,由CO和FLC共同调控[41]。FLC通过扰乱启动子的可及性来负调控SOC1,阻止开花,直到植物重新获得开花能力[42]。SOC1的核心作用是与AP1的同源基因一起调控花序发育的LFY基因[43]。AP1和LFY是将花促进基因连接到花结构发育的两个主要基因[44-45]。SOC1是各个途径调节的交叉点。FLC基因与春化、自主途径密切相关。在春化过程前,FLC抑制花发生,阻止顶端分生组织转化为繁殖结构所必需的生理变化[46]。在长时间暴露于寒冷之后,FLC被抑制,植物开始开花。Michaels和Amasino[47]认为,自主促进与春化途径共同协作使得FLC基因受到抑制。CO基因编码锌指转录因子,其作用受生物钟和光周期的成花素FT调控[48]。
3. 牧草开花的分子机理
3.1 禾本科牧草开花基因研究
3.1.1 黑麦草开花基因研究
多年生黑麦草是世界温带地区普遍种植的优质牧草。Gagic等[17]研究发现多年生黑麦草LpGI基因可能与拟南芥GI基因同源,参与黑麦草的光周期开花时间控制。Skøt等[49]通过关联分析进行候选基因表型的评估,证实LpHd1除对抽穗有促进作用外,还与开花时间显著关联。Fiil等[50]分析多个多年生黑麦草开花控制基因,LpFT3基因的表达由生物钟控制,与日照长度相关,与春化时间密切相关,过表达LpFT3促进开花;LpTFL1与LpFT基因功能相反,抑制黑麦草的开花;LpCO编码的蛋白长时间诱导LpFT的转录,促进多年生黑麦草的开花;LpVRN1春化处理后开始表达,通过抑制VRN2的表达促进植株开花。
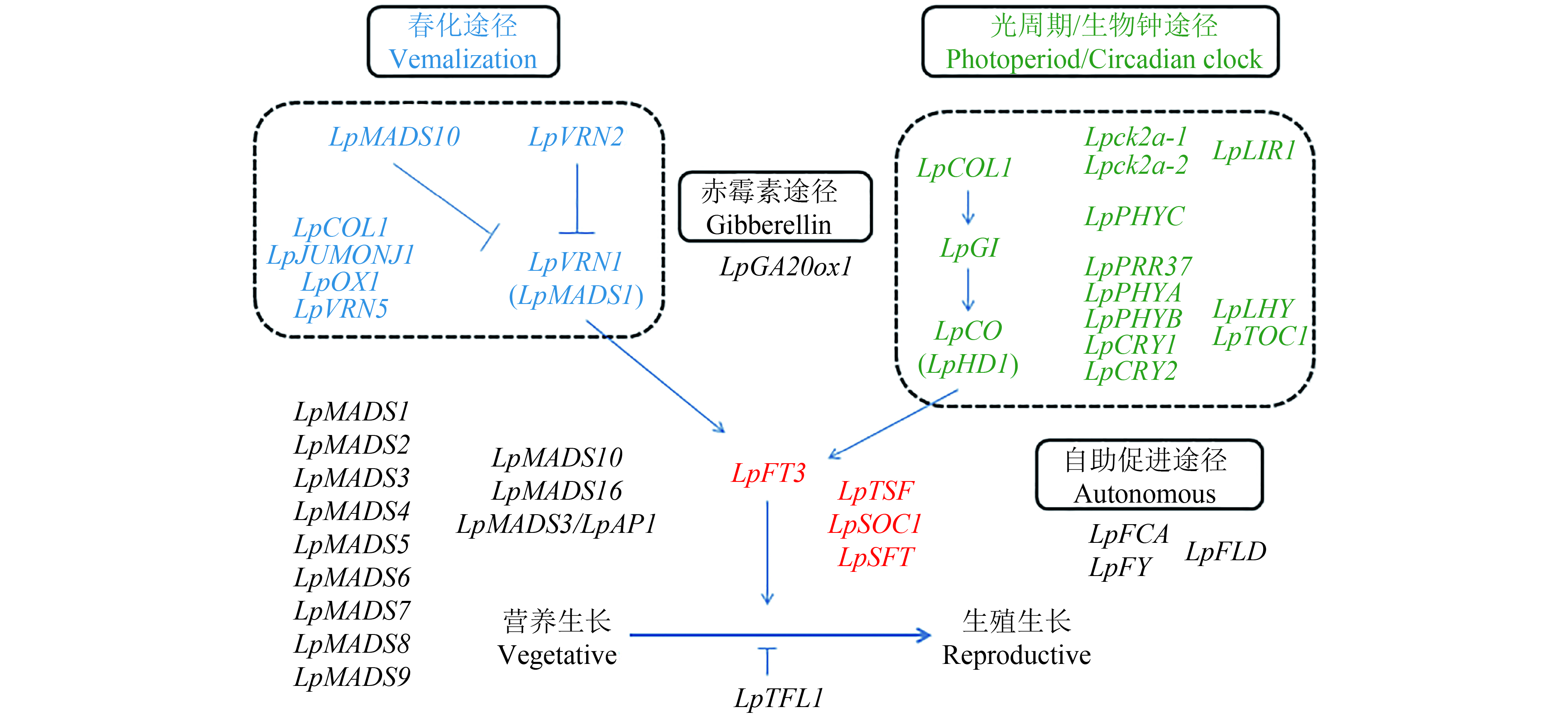
Wang和Forster[51]通过比较基因组学方法鉴定了多年生黑麦草开花诱导途径以及参与的关键基因(图2、表1)。多年生黑麦草的开花时间通常以抽穗期(HD)来衡量,共鉴定了36个与抽穗期相关的QTL,QTL分布在7个连锁群上。多年生黑麦草中鉴定的直系同源基因LpFT3,定位于LG7上,该基因的表达处于生物钟控制之下,与日照长度、春化时间密切相关。第2个中央整合子,SOC1的表达受FT的调控以促进发育,鉴定的SOC1的直系同源基因LpSOC1定位在LG6上。此外,发现MADS-box基因LpMADS1(与LpVRN1相同)和LpMADS2具有SOC1-like基因的功能。春化途径中,LpVRN1是单子叶植物TmVRN1基因的假定直系同源基因,由春化诱导,位于多年生黑麦草的LG4上。3个含MADS-box结构域的基因(LpMADS1、LpMADS2和LpMADS3)被春化处理上调,另一个含MADS-box的基因LpMADS10基于与LpMADS1的蛋白质– 蛋白质相互作用而被确定,并被证明是通过春化处理下调的。LpMADS1与LG4上的LpVRN1基因座相同,LpMADS2、LpMADS3和LpMADS10的位置是未知的。光周期途径中,LpCO基因定位在LG7上,与AtCO具有类似的组织结构,2个外显子和1个含锌指结构域的内含子,启动子区域的序列与水稻OsHD1和大麦HvCO的相应区域也有很高的相似性。LpCOL1位于LG6上,与LG7上报道的其他两个CO-like基因座不同。LpCOL1显示具有与LpCO类似的昼夜节律调节模式,而不影响春化。LG3上的LpGI基因与其他单子叶植物和真核生物的GI基因具有高水平的结构保守性,并表现出昼夜模式。LpGI的组成型表达充分补充了拟南芥、gi-3、突变中的缺陷,证实了黑麦草基因的功能状态。属于自主途径的FLD基因的同源基因也在LG2上被鉴定和定位,但基因功能尚未鉴定。
![]() 图 2 多年生黑麦草开花调控示意图方框内容显示通路名称,箭头表示促进作用,“T”形表示抑制作用。春化途径中的基因以蓝色显示,光周期/生物钟途径中的基因以绿色显示,中央整合基因以红色显示。在花序中具有推定作用的MADS盒基因显示在左下,而来自自主途径的同源基因显示在右下。Figure 2. Perennial ryegrass flowering control schematic diagramPathway names are placed in square boxes. Arrows show induction, “T” show repression. Genes in the vernalisation pathway are shown in blue, genes in photoperiod/circadian clock pathways in green, while central integrator genes are shown in red. MADS-box genes with putative role in floral identity are shown at the bottom left, while gene homologues from the autonomous pathway are shown at the bottom right.表 1 多年生黑麦草开花途径参与基因Table 1. Genes involved in flowering pathway of Perennial ryegrass
图 2 多年生黑麦草开花调控示意图方框内容显示通路名称,箭头表示促进作用,“T”形表示抑制作用。春化途径中的基因以蓝色显示,光周期/生物钟途径中的基因以绿色显示,中央整合基因以红色显示。在花序中具有推定作用的MADS盒基因显示在左下,而来自自主途径的同源基因显示在右下。Figure 2. Perennial ryegrass flowering control schematic diagramPathway names are placed in square boxes. Arrows show induction, “T” show repression. Genes in the vernalisation pathway are shown in blue, genes in photoperiod/circadian clock pathways in green, while central integrator genes are shown in red. MADS-box genes with putative role in floral identity are shown at the bottom left, while gene homologues from the autonomous pathway are shown at the bottom right.表 1 多年生黑麦草开花途径参与基因Table 1. Genes involved in flowering pathway of Perennial ryegrass基因名 Gene name 基因编号 GeneBank accession 同源基因 Homologous gene 通路/功能 Pathways/Function LpTFL1 AF316419 HvTFL1 抑制 Repressor LpSOC1 AtSOC1 中央整合子 Central integrator LpFT3 DQ309592 AtFT、OsHd3a、OsFTL2、HvFT、TaFT1 LpTSF AtFT LpSFT HvFT4 LpPRR37 OsPRR37 光周期 Photoperiod LpPRR73 OsPRR73 LpGI DQ534010 AtGI、HvGI、TaGI、OsGI LpPHYA AtPHYA LpPHYB AtPHYB LpPHYC PHYC LpCRY1 AtCRY1 LpCRY2 AtCRY2 LpTOC1 TOC1 LpCO AY600919 AtCO,OsHd1、HvCO1、TaHd1-1 (LpHD1) AM489608 Lpck2a-2 AB213317 Wheat tck2a 生物钟 Circadian clock Lpck2a-1 AB213316 OsCK2a LpCOL1 DQ145928 AtCO、OsHd1 LpLHY LHY LpLIR1 DQ206624 rice LIR LpGA20ox1 DQ071620 赤霉素 Gibberellin LpFLD FLD 自主促进 Autonomous LpFCA AY654582 AtFCA LpFY AtFY LpVrn2-2 DQ202716 TmVrn2 春化 Vernalization LpOX1 DQ145927 2OG-Fe(II) oxygenase LpJMJC JUMONJI-like LpVRN1 AY198326 TmVrn1、OsAP1、AtAP1、LtMADS1 (LpMADS1) GQ258851 LpVRN5 VRN5 LpMADS2 AY198327 AtAP1、LtMADS2 LpMADS3 AY198328 AtAP1、barley BM3 LpMADS4 AY198329 TaMADS12 花发育基因 Flower development LpMADS5 AY198330 barley BM9 LpMADS6 AY198331 OsMADS5 花发育基因 Flower development LpMADS7 AY198332 OsE31254 LpMADS8 AY198333 OsMADS24 LpMADS9 AY198334 Barley BM7 LpMADS10 DQ110009 OsMADS22、VRT2、SVP/AGL24-like 春化分生组织特性基因
Vernalization meristem identityLpMADS16 DQ110011 SVP-like 3.1.2 鸭茅开花基因研究
鸭茅(Dactylis glomerata)是禾本科鸭茅属的多年生牧草,在世界温带地区普遍分布和种植。近年来,鸭茅开花分子的探索得到了很好的开展。Xie 等[52]以晚熟和早熟鸭茅基因型为亲本构建了F2作图群体,共检测到7个影响开花性状的QTL,分别位于第2、5、6组,解释表型变异在7.85%~24.19%。基于水稻、小麦、黑麦草、高羊茅(Festuca elata)等物种的开花基因(如FT、Hd、VRN等)的保守序列设计基因探针,对鸭茅开花基因序列进行捕获,涉及引物在作图群体里扩增,最终在鸭茅第3号连锁群上成功定位了VRN3基因[53]。Zhao 等[54]对鸭茅F2 群体的214个单株和亲本进行高通量测序,构建了四倍体高密度遗传连锁图谱,结合两年三点的开花表型数据,定位了与开花相关的CO、FT及VRN1基因位点5个。Zhao等[55]利用CONSTANS (DgCO1),FLOWERING TIME (DgFT1)、a VRN1 like MADS-box (DgMADS)和PHOTOPERIOD (DgPPD1-like)共4个开花候选基因在150个单株组成的关联群体里进行关联分析,研究发现DgCO1基因与鸭茅开花显著关联。
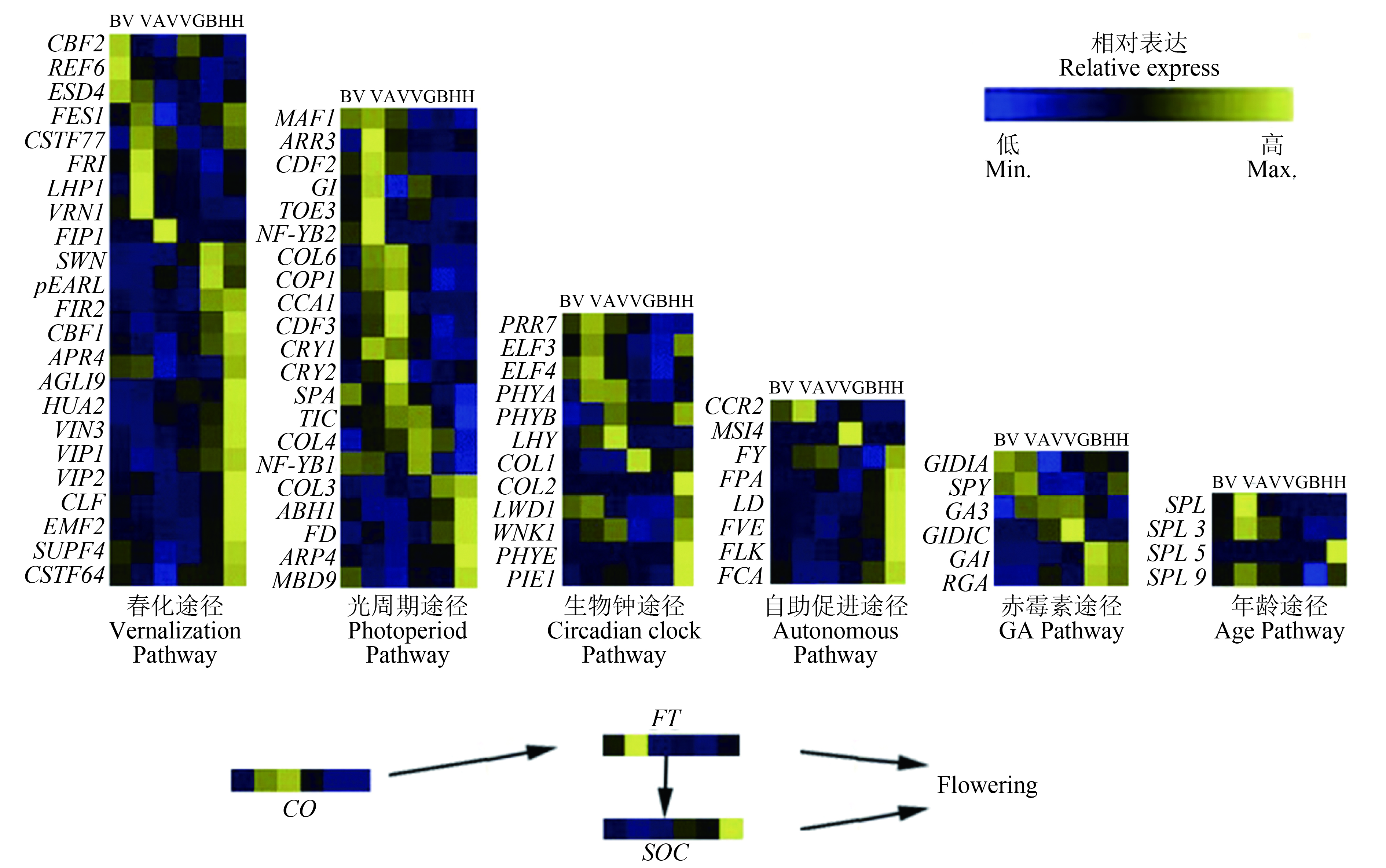
Feng等[56]利用鸭茅不同时期花序进行转录组,测序共鉴定了77个与开花相关的基因(图3)。其中,有VRN1、VIN3和FRI等24个与春化有关的候选基因,TIC、COL和FD等20个与光周期途径有关的候选基因,以及12个生物钟候选基因、8个自主促进候选基因、5个赤霉素候选基因、4个衰老候选基因和4个整合基因。光周期途径相关基因在第2阶段和第5阶段富集,表明该途径中的基因可能对不同长度的日照时间有反应。此外,光合作用相关基因在春化期表现出强烈的阶段依赖性和高表达水平,这可能表明这些基因是紧密共表达的。VRN1和FRI等大多数春化相关基因在第2阶段和第5阶段高水平表达。VRN1在第2阶段的高水平表达符合低温诱导VRN1。在拟南芥中,FRI是一个上游调控因子,通过激活FLC,抑制整合子FT的表达,从而导致晚开花。数据显示,在第2阶段期间FRI高表达,然后在第3阶段期间急剧下降,表明春化导致FRI降解。FLC在春化阶段表达量较低,随后逐渐增加,直至抽穗期。相反,FT在春化过程中表达量较高,而在其他时期表达量较低,这说明低温抑制FLC表达和解除FT抑制导致开花。
![]() 图 3 鸭茅开花调控示意图热图显示候选基因在不同阶段的相对表达,y轴代表已确定的候选基因,而x轴显示不同阶段。BV,春化前;V,春化期;AV,春化后;VG,营养生长期;BH,抽穗前;H,抽穗期。从BV到V定义为阶段1(P1),V到AV定义为第1阶段(P2),AV到VG定义为第2阶段(P3),VG到BH定义为第3阶段4(P4), BH到H定义为第5阶段(P5)。Figure 3. Dactylis glomerata flowering control schematic diagramThe heat map shows relative expression of candidate genes at different stages. The y-axis represents identified candidate genes and the x-axis shows the different stages. BV, before vernalization; V, vernalization; AV, after vernalization; VG, vegetative growth; BH, before heading; H, heading. From BV to V is defined as phase 1 (P1), V to AV is defined as phase 2 (P2), AV to VG is defined as phase 3 (P3), VG to BH is defined as phase 4 (P4), and BH to H is defined as phase 5 (P5).
图 3 鸭茅开花调控示意图热图显示候选基因在不同阶段的相对表达,y轴代表已确定的候选基因,而x轴显示不同阶段。BV,春化前;V,春化期;AV,春化后;VG,营养生长期;BH,抽穗前;H,抽穗期。从BV到V定义为阶段1(P1),V到AV定义为第1阶段(P2),AV到VG定义为第2阶段(P3),VG到BH定义为第3阶段4(P4), BH到H定义为第5阶段(P5)。Figure 3. Dactylis glomerata flowering control schematic diagramThe heat map shows relative expression of candidate genes at different stages. The y-axis represents identified candidate genes and the x-axis shows the different stages. BV, before vernalization; V, vernalization; AV, after vernalization; VG, vegetative growth; BH, before heading; H, heading. From BV to V is defined as phase 1 (P1), V to AV is defined as phase 2 (P2), AV to VG is defined as phase 3 (P3), VG to BH is defined as phase 4 (P4), and BH to H is defined as phase 5 (P5).3.1.3 羊茅属牧草开花基因研究
陈锡等[57]通过RACE技术,用高羊茅“黔草1号”作为材料,克隆植物开花基因FT,命名为FaFT2。推断FaFT2与高羊茅光周期具有一定的关联性,过度表达可能导致植株提前开花(表2)。王小利等[58]根据多年生黑麦草VRN1 基因序列设计的引物,用高羊茅茎基部组织的mRNA 为模板,进行RT-PCR 分析。结合3' RACE 和5' RACE 法从高羊茅中扩增出VRN1 的全长cDNA 序列,命名为FaVRN1,发现该基因具有高度的保守性,是开花诱导基因。
表 2 牧草开花基因Table 2. Forage grass flowering gene草种
Species开花基因
Flowering gene基因功能
Gene function参考文献
Reference多年生黑麦草
Lolium perenneLpFT3 由生物钟控制表达,与日长、春化时间相关,过度表达促进开花
Controlled by the circadian clock, related to the day length and the vernalisation time, over expression promotes flowering.[51] LpCO 长时间诱导,编码的蛋白促进开花
It is induced by long days and the encoded protein promotes flowering.[50] LpTFL1 与 FT 基因功能相反,参与开花抑制
In contrast to FT, it is involved in repression of flowering.[50] LpVRN1 春化诱导,表达模式与开花时间变化密切相关
It is induced by vernalisation and the expression pattern is correlated with variation in flowering time.[51] LpGI 与拟南芥 GI 同源,参与光周期开花时间控制
It is orthologous to Arabidopsis GI and involved in photoperiodic flowering time control.[17] LpHd1 水稻 OsHd1 的直系同源基因,影响开花时间
An ortholog of rice OsHd1 in affecting time of flowering.[49] 高羊茅
Festuca arundinaceaFaFT2 受光照调控,过表达促进开花
Controlled by light, over expression promotes flowering.[57] FaVRN1 开花诱导基因
Flowering inducing gene.[58] 草甸羊茅
Festuca pratensisFpVRN1 诱导 FT1 基因,在春季促进生殖器官的发育
FT1 gene expression is induced to promote development of reproduction organs in spring.[59] FpCO 直接结合 FT 基因的启动子以激活表达
Directly bind to the promoter of the FTgene to activate expression.FpFT1 促进开花
Promote floweringFpCK2α 在短日照条件下通过间接相互作用,增强 CO-like 蛋白质的功能
Enhances function of the CO-like protein through indirect interaction under short-day.扁穗冰草
Agropyron cristatumACOL CO 候选基因,在叶片中特异性表达
It is a CONSTANS (CO) candidate gene and showed specific expression in leaves.[60] 二穗短柄草
Brachypodium distachyonEZL1 拟南芥 CURLY LEAF 1 的直系同源基因,影响开花时间
An ortholog of Arabidopsis CURLY LEAF 1 in affecting flowering time.[61] 紫花苜蓿
Medicago sativaSPL13 调控开花时间和营养生长
It is regulating flowering time and vegetative growth.[62] miR156 过表达导致生物量产量加倍,开花时间延迟,纤维素含量增加和木质素含量减少Over expression resulted in a twofold increase in biomass yield, delayed flowering time, enhanced cellulose content, and reduced lignin. [63] 蒺藜苜蓿
Medicago truncatulaSTENOFOLIA(STF) 参与植株叶片的扩展,增加叶绿素含量,促进开花
It is involved in leaf expansion, increased chlorophyll content and accelerated flowering.[64] 红三叶
Trifolium pratenseWAT1 降低结实能力,减少种子产量
Reduces firmness and seed yield.[65] NIP4-1 提高种子产量
Increases seed yield.ZFP4 与结实相关
Closely related to seed setting.ERF106 控制种子重量
Controls seed weight.Pt4 提高种子产量
Increases seed yield.地三叶
Trifolium subterraneumCO-like 和 WNK5-like 蛋白介导远红光下的开花启动
Floral initiation under FR-enriched light is mediated by CO-like and WNK5-like proteins.[66] 白羽扁豆
Lupinus albus显性基因 Ef1 和 Ef2 当(Ef1_,Ef2_)同时出现时,表型早开花;(ef1ef1, ef2ef2)同时缺失时,表型晚开花;如果只有一个基因(Ef1_,ef2ef2 or ef1ef1, Ef2_)存在时,表型为中间体
When both dominant genes were present (Ef1_, Ef2_) the phenotype was early flowering, when both dominant genes were absent (ef1ef1, ef2ef2) the phenotype was late flowering and if only one gene was present (Ef1_, ef2ef2 or ef1ef1, Ef2_), the phenotype was intermediate.[67] 在草甸羊茅(Festuca pratensis)研究中,Shinozuka等[59]评估了开花时间相关基因的核苷酸多样性。对草甸羊茅VRN1、CO、FT1和CK2α直系同源基因FpVRN1、FpCO、FpFT1和FpCK2α进行重新测序,并对3个样本组内的核苷酸多样性进行评估。证明FpVRN1诱导FT1基因表达,在春季促进生殖器官的发育;FpCO联合FT基因的启动子以启动表达;FpFT1诱导植株提前开花;FpCK2α在短日照条件下通过间接相互作用,增强CO-like蛋白质的功能。
3.1.4 扁穗冰草开花基因研究
Zeng等[60]在扁穗冰草(Agropyron cristatum)研究中通过RNA-Seq和差异基因表达分析,研究开花发育的基因表达。对四倍体扁穗冰草的3个发育阶段,即茎伸长期(VS)、孕穗期(BS)和开花期(AS)进行转录组分析。总共确定了113个开花时间相关基因,123个MADS-box基因和22个CONSTANS-Like(COL)基因,其中发现ACOL作为CO候选基因,在叶片中特异性表达。产生一套新的基因组资源,用于鉴定和表征与扁穗冰草开花相关的基因和途径。该发现不仅证明RNA-Seq和差异基因表达分析对无参考基因组物种的复杂性状的相关基因的有效性,而且提高了我们对扁穗冰草光周期– 生物钟– CO– FT的开花启动调控途径的认识。
3.1.5 二穗短柄草开花基因研究
二穗短柄草(Brachypodium distachyon)属禾本科早熟禾亚科,由于其为一年生,生命周期短;株型小、易栽培;天然状态下自花授粉;易遗传转化;具有丰富的遗传变异材料。因此是研究麦类植物基因组学的优良模式材料。Lomax等[61]在二穗短柄草中对减少春化需求的突变体进行筛选时,鉴定出了隐性等位基因ENHANCER OF ZESTE-LIKE 1(EZL1)。EZL1是拟南芥CURLY LEAF 1的直系同源基因,是编码PRC2的催化亚基。在EZL1突变体的转录组研究中,发现相比于无春化的野生型材料,EZL1-1中有近1 400个基因差异表达,并且约有800个基因在EZL1-1突变体中上调。其中前十个最高表达的基因中,有4个涉及开花:AGAMOUS (AG)、VRN1、SOC1和SEPALLATA 3 (SEP3)。当AG、VRN1、SOC1和SEP3过度表达时,可能会促进EZL1突变体表型快速开花。
3.2 豆科牧草开花基因研究
3.2.1 苜蓿属牧草开花基因研究
Gao等[62]在紫花苜蓿(Medicago sativa)研究中表明,存在于植物分生组织中的SPL13在营养生长和生殖发育过程中起着至关重要的作用。通过SPL13过表达和SPL13基因沉默两种紫花苜蓿植株进行基因功能研究,过表达植株生长阻滞、枝条扭曲、叶片向上卷曲;沉默植株观察到更多的侧枝和开花时间延迟,表明SPL13参与枝梢发育的调控,过表达促进开花。使用基于新一代测序技术的SPL13 RNAi植物的转录组分析来鉴定可能被SPL13直接靶向和下调的下游基因,将SPL13与MYB112基因联系起来,表明MYB112受到SPL13的靶向和下调,基于下一代测序的转录组分析和染色质免疫沉淀分析,表明MYB112可能参与调控苜蓿营养生长的过程。Aung等[63]研究表明,过度表达microRNA156 (miR156),紫花苜蓿表现出一种明显的表型,节间长度缩短,开花时间延缓,纤维素含量增加,木质素含量减少,生物量产量明显提高。
在蒺藜苜蓿(Medicago truncatula)研究中,WOX家族成员STENOFOLIA(STF)在叶片的背面和正面交叉处的特定叶细胞中表达。Liu等[64]通过在六倍体小麦中表达M. truncatula STF,与对照相比,转基因植物明显呈现叶片加宽,开花加速和叶绿素含量增加,表明STF参与植株叶片的扩展。
3.2.2 三叶草属开花基因研究
在红三叶(Trifolium pratense)的研究中,Kovi等[65]选择对具有弱结实率的‘Tripo’品种和具有强结实率的‘Lasang’品种的基因型进行转录组分析。发现WAT1相关蛋白是主要负责跨膜转运蛋白活性的细胞壁蛋白,该基因在‘Tripo’花蕾中的早花和中花发育时期表达下调,对其结实能力和种子产量都有负面影响;NIP4-1是水通道蛋白基因家族成员,在‘Lasang’中的早花和中花阶段显著上调,提高该品种的种子产量;ZFP4在早花和中花阶段高度表达,增强结实;ERF106是APETALA2(AP2)基因家族一部分,在‘Lasang’花蕾的早花和中花阶段过度表达,在‘Tripo’后花阶段表达下调,控制种子重量;Pt4属于调控植物细胞磷稳态的基因,在中花和后花阶段过表达,提高种子产量。在红三叶品种中对上述基因研究表明,这些基因均在不同开花时期出现了基因表达上的差异,进而对种子的结实能力以及种子的产量产生影响。
Navarro等[66]使用深度测序技术调查长日照植物地三叶(Trifolium subterraneum)在3个时间点的不同光谱下的转录活性,发现以地三叶为模型的长日照植物在FR光下会加速与开花途径相关基因的上调,结果表明COL和WNK5-like蛋白介导富含FR光下的开花启动。
3.2.3 白羽扁豆开花基因研究
Adhikari等[67]通过检测白羽扁豆(Lupinus albus)F1、F2和F2衍生的F3代,在两个育种群体中研究开花时间的遗传,发现白羽扁豆开花受两个互补的显性基因Ef1和Ef2控制:当两个显性基因都存在时(Ef1_、Ef2_),表型为早花;当两个显性基因都不存在时(ef1ef1、ef2ef2),表型延迟开花;如果只有一个基因存在(Ef1_、ef2ef2或ef1ef1、Ef2_),表型是中等的。
4. 展望
开花是牧草重要的生命过程,是众多基因控制的数量性状。其调控对于牧草生存和繁殖至关重要,受到环境条件和发育规律的影响,这种影响牵涉复杂的网络信号。因此,牧草开花的分子机理研究对控制牧草的开花进程起重要作用,同时可以有效解决牧草产量低、质量差和经济效益低下等问题,进而培育优良品种,实现牧草的高产和稳产。目前,随着分子生物学的发展,人们对于牧草开花机制进行了大量的研究,已形成逐步完熟的开花体系,但仍有问题尚未解决。
首先,目前对于开花调控分子机理的关注主要集中在拟南芥、水稻、小麦等模式植物中,对多年生牧草的开花研究很少,所以当前植物界开花机制的适用性还需进一步深入。
其次,尽管开花特异性信号传导途径的基因功能在非模型牧草中是非常保守的,但是这些基因的功能特异性有时是不同的。有些基因并非在相同的植物物种中具有相同的功能,其基因功能可能根据环境条件特别是季节长度的纬度变化而发生改变。
此外,虽然FT基因调控牧草开花的研究越来越深入,但其作用机制仍存在很多问题。FT基因如何调控下游基因,FT在开花途径交汇节点如何整合信号等问题都需进一步探索。为研究FT转运机制,人们提出很多方法。其中筛选FT互作蛋白法有助于进一步研究FT调控开花的分子机理,同时有助于发掘FT蛋白调控其他生长发育的功能;GI基因参与多条开花途径,并与其他基因相互作用,但是其确切的分子功能仍不清楚,如何受昼夜节律调控,如何控制CO的表达等都需进一步深入研究。对草类植物MADS-box基因的研究很少,因此有必要继续扩大研究范围以补充MADS-box基因的作用机制。
如果以上问题可以解决,牧草开花作用机制的研究将会更加透彻。随着测序技术的进步和生物信息学的发展,将会有更多的遗传信息被发现,这对牧草培育具有重要意义。
参考文献
[1] WOODS D P, MCKEOWN M A, DONG Y, PRESTON J C, AMASINO R M. Evolution of VRN2/GhD7-like genes in vernalization-mediated repression of grass flowering. Plant Physiology, 2016, 170(4): 2124-2135. doi: 10.1104/pp.15.01279
[2] DENG W, CASAO M C, WANG P, SATO K, HAYES P M, FINNEGAN E J, TREVASKIS B. Direct links between the vernalization response and other key traits of cereal crops. Nature Communication, 2015, 6(6): 5882.
[3] VOGEL K P, BREJDA J J, WALTERS D T, BUXTON D R. Switchgrass biomass production in the midwest USA. Agronomy Journal, 2002, 94(3): 413-420. doi: 10.2134/agronj2002.0413
[4] MOURADOV A, CREMER F, COUPLAND G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell, 2002, 14(Suppl): S111-S130.
[5] BOSS P K, BASTOW R M, MYLNE J S, DEAN C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell, 2004, 16(Suppl): S18-S31.
[6] PENG F Y, HU Z, YANG R C. Bioinformatic prediction of transcription factor binding sites at promoter regions of genes for photoperiod and vernalization responses in model and temperate cereal plants. BMC Genomics, 2016, 17(1): 573. doi: 10.1186/s12864-016-2916-7
[7] DORCA-FORNELL C, GREGIS V, GRANDI V, COUPLAND G, COLOMBO L, KATER M M. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant Journal for Cell & Molecular Biology, 2011, 67(6): 1006-1017.
[8] COCKRAM J, JONES H, LEIGH F J, O'SULLIVAN D, POWELL W, LAURIE D A, GREENLAND A J. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. Journal of Experimental Botany, 2007, 58(6): 1231-1244. doi: 10.1093/jxb/erm042
[9] HIGGINS J A, BAILEY P C, LAURIE D A. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One, 2010, 5(4): e10065. doi: 10.1371/journal.pone.0010065
[10] CHIRUTA C, FILIPOV F, CALIN M. Estimating the duration of daylight in a given time of the year depending on the latitude of the location. Research Journal of Agricultural Science, 2010, 42(3): 71-76.
[11] WADA M, SHIMAZAKI K, IINO M. Light Sensing in Plants. 2005. Berlin: Springer, 2005: 333-337.
[12] FERNÁNDEZ V, TAKAHASHI Y, LEGOURRIEREC J, COUPLAND G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. The Plant Journal, 2016, 86(5): 426-440.
[13] ONOUCHI H, IGEÑO M I, PÉRILLEUX C, GRAVES K, COUPLAND G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell, 2000, 12(6): 885-900. doi: 10.1105/tpc.12.6.885
[14] IZAWA T, MIHARA M, SUZUKI Y, GUPTA M, ITOH H, ATSUSHI J, NAGANO, MOTOYAMA R, SAWADA Y, YANO M, MASAMI YOKOTA HIRAI, MAKINO A, NAGAMURA Y. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell, 2011, 23(5): 1741-1755. doi: 10.1105/tpc.111.083238
[15] BENDIX C, MENDOZA J M, STANLEY D N, MEELEY R, HARMON F G. The circadian clock-associated gene gigantea1, affects maize developmental transitions. Plant Cell & Environment, 2013, 36(7): 1379-1390.
[16] MARTIN J, STORGAARD M, ANDERSEN C H, NIELSEN K K. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Molecular Biology, 2004, 56(2): 159-169. doi: 10.1007/s11103-004-2647-z
[17] GAGIC M, FAVILLE M, KARDAILSKY I, PUTTERILL J. Comparative genomics and functional characterisation of the GIGANTEA, gene from the temperate forage perennial ryegrass Lolium perenne. Plant Molecular Biology Reporter, 2015, 33(4): 1-9.
[18] HECHT V, LAURIE R E, WELLER J. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Frontiers in Plant Science, 2014, 5(17): 486.
[19] 王鹏, 张春庆, 陈化榜, 吴承来. 小麦冬性强弱评价体系的建立. 生态学报, 2012, 32(4): 1230-1240. WANG P, ZHANG C Q, CHEN H B, WU C L. The evaluation system of strength of winterness in wheat. Acta Ecologica Sinica, 2012, 32(4): 1230-1240.
[20] ZHANG X K, XIAO Y G, ZHANG Y, XIA X C, DUBCOVSKY J, HE Z H. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Science, 2008, 42(2): 1690-1694.
[21] 安艳荣. 二穗短柄草VILs基因对开花时间的调控机理研究. 青岛: 山东农业大学博士学位论文, 2015. AN Y R. Mechanism of regulation on flowering time by VIL genes in Brachypodium distachyon. PhD Thesis. Qingdao: Shandong Agricultural University, 2015.
[22] YOSHIDA T, NISHIDA H, ZHU J, NITCHER R, DISTELFELD A, AKASHI Y, KATO K, DUBCOVSKY J. Vrn-D4, is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theoretical & Applied Genetics, 2010, 120(3): 543-552.
[23] PIDAL B, YAN L, FU D, ZHANG, TRANQUILLI G, DUBCOVSKY J. The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. Journal of Heredity, 2009, 100(3): 355-364. doi: 10.1093/jhered/esp002
[24] DUBCOVSKY J, CHEN C, YAN L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Molecular Breeding, 2005, 15(4): 395-407. doi: 10.1007/s11032-005-0084-6
[25] YAN L, LOUKOIANOV A, BLECHL A, TRANQUILLI G, RAMAKRISHNA W, SANMIGUEL P, BENNETZEN J L, ECHENIQUE V, DUBCOVSKY J. The wheat VRN2 gene is a flowering repressor downregulated by vernalization. Science, 2004, 303(5664): 1640-1644. doi: 10.1126/science.1094305
[26] KIPPES N, ZHU J, CHEN A, VANZETTI L, LUKASZEWSKI A, NISHIDA H, KATO K, DVORAK J, DUBCOVSKY J. Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Molecular Genetics and Genomics, 2014, 289(1): 47-62. doi: 10.1007/s00438-013-0788-y
[27] MARQUARDT S, BOSS P K, HADFIELD J, DEAN C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. Journal of Experimental Botany, 2006, 57(13): 3379-3386. doi: 10.1093/jxb/erl073
[28] CHENG J Z, ZHOU Y P, LV T X, XIE C P, TIAN C E. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiology & Molecular Biology of Plants, 2017, 23(3): 1-9.
[29] SIMPSON G G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology, 2004, 7(5): 570-574. doi: 10.1016/j.pbi.2004.07.002
[30] EOM H, PARK S J, KIM M K, KIM H, KANG H, LEE I. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant Journal for Cell & Molecular Biology, 2017, 93(1): 79-91.
[31] YABUTA T, HAYASI T. Biochemical studies of 'bakanae' fungus of rice. Journal of the Imperial Agricultural Experimental Station Nisigahara Tokyo, 1940, 25(3): 365-400.
[32] 黄桃鹏, 李媚娟, 王睿, 李玲. 赤霉素生物合成及信号转导途径研究进展. 植物生理学报, 2015(8): 1241-1247. HUANG T P, LI M J, WANG R, LI L. Progress in study of gibberellins biosynthesis and signaling transduction pathway. Plant Physiology Journal, 2015(8): 1241-1247.
[33] BLAZQUEZ M A, GREEN R, NILSSON O, SUSSMAN M R, WEIGEL D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell, 1998, 10(5): 791-800. doi: 10.1105/tpc.10.5.791
[34] XUE L J, ZHANG J J, XUE H W. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PloS One, 2012, 7(2): e31081. doi: 10.1371/journal.pone.0031081
[35] 黄磊玉. 植物开花基因的研究进展. 农家科技, 2017(3): 257. HUANG L Y. Research progress of flowering genes in plants. Nongjia Keji, 2017(3): 257.
[36] 李昱, 罗志鹏, 赵淑清. 拟南芥开花时间调控的整合途径. 植物生理学报, 2007, 43(5): 799-804. LI Y, LUO Z P, ZHAO S Q. Integration pathway of flowering time control in Arabidopsis. Plant Physiology Journal, 2007, 43(5): 799-804.
[37] 袁敏, 邢朝斌, 葛伟娜, 王莉, 郭棣. 拟南芥开花诱导基因FT的蛋白表达及纯化. 基因组学与应用生物学, 2017, 36(8): 3053-3056. YUAN M, XING C B, GE W N, WANG L, GUO D. Protein expression and purification of flowering inducer gene FT in Arabidopsis thaliana. Genomics and Applied Biology, 2017, 36(8): 3053-3056.
[38] 兰树斌, 李建国. 植物LFY基因的研究进展. 基因组学与应用生物学, 2007, 26(s1): 132-137. LAN S B, LI J G. Advance of studies on LFY gene in plants. Genomics and Applied Biology, 2007, 26(s1): 132-137.
[39] 王利琳, 梁海曼, 庞基良, 朱睦元. 拟南芥LEAFY基因在花发育中的网络调控及其生物学功能. 遗传, 2004, 26(1): 137-142. doi: 10.3321/j.issn:0253-9772.2004.01.026 WANG L L, LIANG H M, PANG J L, ZHU M Y. Regulation network and biological roles of LEAFY in Arabidopsis thaliana in floral development. Hereditas, 2004, 26(1): 137-142. doi: 10.3321/j.issn:0253-9772.2004.01.026
[40] SAMACH A, ONOUCHI H, GOLD S E, DITTA G S, SCHWARZ-SOMMER Z, YANOFSKY M F, COUPLAND G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science, 2000, 288: 1613-1616. doi: 10.1126/science.288.5471.1613
[41] HEPWORTH S R, VALVERDE F, RAVENSCROFT D, MOURADOV A, COUPLAND G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo Journal, 2002, 21(16): 4327-4337. doi: 10.1093/emboj/cdf432
[42] SEARLE I, HE Y, TURCK F, VINCENT C, FORNARA F, KRÖBER S, AMASINO R A, COUPLAND G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development, 2006, 20(7): 898-912.
[43] SCHULTZ E A, HAUGHN G W. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell, 1991, 3(8): 771-781. doi: 10.1105/tpc.3.8.771
[44] LOHMANN J U, HONG R L, HOBE M, BUSCH M A, PARCY F, SIMON R, WEIGEL D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell, 2001, 105(6): 793-803. doi: 10.1016/S0092-8674(01)00384-1
[45] GUSTAFSON-BROWN C, SAVIDGE B, YANOFSKY M F. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell, 1994, 76(1): 131-143. doi: 10.1016/0092-8674(94)90178-3
[46] SHELDON C C, ROUSE D T, FINNEGAN E J, PEACOCK W J, DENNIS E S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C(FLC). Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(7): 3753-3758. doi: 10.1073/pnas.97.7.3753
[47] MICHAELS S D, AMASINO R M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 1999, 11(5): 949-956. doi: 10.1105/tpc.11.5.949
[48] PUTTERILL J, ROBSON F, LEE K, SIMON R, COUPLAND G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell, 1995, 80(6): 847-857. doi: 10.1016/0092-8674(95)90288-0
[49] SKØT L, SANDERSON R, THOMAS A, SKØT K, THOROGOOD D, LATYPOVA G, ASP T, ARMSTEAD I. Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiology, 2011, 155(2): 1013-1022. doi: 10.1104/pp.110.169870
[50] FIIL A, LENK I, PETERSEN K, JENSEN C S, NIELSEN K K, SCHEJBEL B, ANDERSEN J R, LÜBBERSTEDT T. Nucleotide diversity and linkage disequilibrium of nine genes with putative effects on flowering time in perennial ryegrass (Lolium perenne L.). Plant Science, 2011, 180(2): 228-237. doi: 10.1016/j.plantsci.2010.08.015
[51] WANG J, FORSTER J W. Flowering time regulation in perennial ryegrass. Euphytica, 2017, 213(5): 106. doi: 10.1007/s10681-017-1896-2
[52] XIE W G, ROBINS J G, BUSHMAN B S. A genetic linkage map of tetraploid orchardgrass (Dactylis glomerata L.) and quantitative trait loci for heading date. Genome, 2012, 55(5): 360-369. doi: 10.1139/g2012-026
[53] 谢文刚. 鸭茅分子遗传连锁图谱构建及开花基因定位. 成都: 四川农业大学博士学位论文, 2013. XIE W G. Genetic linkage map and flowering time gene mapping in orchardgrass (Dactylis glomerata L.). PhD Thesis. Chengdu: Sichuan Agricultural University, 2013.
[54] ZHAO X X, HUANG L K, ZHANG X Q, WANG J P, YAN D F, LI J, TANG L, LI X L, SHI T W. Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq. Scientific Reports, 2016, 6: 29345. doi: 10.1038/srep29345
[55] ZHAO X X, BUSHMAN B S, ZHANG X Q, ROBBINS M D, LARSON S R, ROBINS J G, THOMAS A. Association of candidate genes with heading date in a diverse Dactylis glomerata population. Plant Science, 2017, 265: 146-153.
[56] FENG G, HUANG L, LI J, WANG J, XU L, PAN L, ZHAO X, WANG X, HUANG T, ZHANG X. Comprehensive transcriptome analysis reveals distinct regulatory programs during vernalization and floral bud development of orchardgrass (Dactylis glomerata L.). BMC Plant Biology, 2017, 17(1): 216. doi: 10.1186/s12870-017-1170-8
[57] 陈锡, 赵德刚, 陈莹, 李小冬, 吴佳海, 王小利. 高羊茅FaFT2基因克隆及表达分析. 植物生理学报, 2017(8): 1523-1531. CHEN X, ZHAO D G, CHEN Y, LI X D, WU J H, WANG X L. Cloning and expression analysis of FaFT2 gene in tall fescue. Plant Physiology Journal, 2017(8): 1523-1531.
[58] 王小利, 陈伟, 李晚忱, 吴佳海, 刘晓霞, 杨义成. 高羊茅春化基因FaVRN1的克隆与分析. 核农学报, 2009, 23(5): 778-784. WANG X L, CHEN W, LI W C, WU J H, LIU X X, YANG Y C. Cloning and characterization of vernalizational gene FaVRN1 from tall fescue. Journal of Nuclear Agricultural Sciences, 2009, 23(5): 778-784.
[59] SHINOZUKA H, HAND M L, COGAN N O, SPANGENBERG G C, FORSTER J W. Nucleotide diversity of vernalization and flowering-time-related genes in a germplasm collection of meadow fescue (Festuca pratensis Huds.) Darbysh.). Ecology & Evolution, 2013, 3(13): 4415-4426.
[60] ZENG F, BILIGETU B, COULMAN B, SCHELLENBERG M P, FU Y B. RNA-Seq analysis of gene expression for floral development in crested wheatgrass (Agropyron cristatum L.). PLoS One, 2017, 12(5): e0177417. doi: 10.1371/journal.pone.0177417
[61] LOMAX A, WOODS D P, DONG Y, BOUCHÉ F, RONG Y, MAYER K S, ZHONG X, AMASINO R M. An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon. Plant Journal for Cell & Molecular Biology, 2018, 93(5): 871-882.
[62] GAO R, GRUBER M Y, AMYOT L, HANNOUFA A. SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Molecular Biology, 2018, 96(1/2): 1-15. doi: 10.1007/s11103-017-0689-2
[63] AUNG B, GRUBER M Y, HANNOUFA A. The MicroRNA156 system: A tool in plant biotechnology. Biocatalysis & Agricultural Biotechnology, 2015, 4(4): 432-442.
[64] LIU M, LEI L, MIAO F, POWERS C, ZHANG X, DENG J, TADEGE M, CARVER B F, YAN L. The STENOFOLIA gene from Medicago alters leaf width, flowering time and chlorophyll content in transgenic wheat. Plant Biotechnology Journal, 2018, 16(1): 186-196. doi: 10.1111/pbi.2018.16.issue-1
[65] KOVI M R, AMDAHL H, ALSHEIKH M, ROGNLI O A. De novo and reference transcriptome assembly of transcripts expressed during flowering provide insight into seed setting in tetraploid red clover. Scientific Reports, 2017, 7: 44383. doi: 10.1038/srep44383
[66] NAVARRO M P, RIBALTA F M, HURGOBIN B, CROSER J S, KAUR P. Gene networks underlying faster flowering induction in response to far-red light. BioRxiv, 2017.
[67] ADHIKARI K, BUIRCHELL B, YAN G, SWEETINGHAM M. Two complementary dominant genes control flowering time in albus lupin(Lupinus albus L.). Plant Breeding, 2011, 130(4): 496-499. doi: 10.1111/pbr.2011.130.issue-4
-
图 1 冬小麦VRN1、VRN2和VRN3反馈调节环
图1显示光周期敏感的冬小麦中VRN1、VRN2和VRN3基因之间的相互作用,箭头表示促进作用,“T”形表示抑制作用。
Figure 1. Winter wheat VRN1, VRN2 and VRN3 feedback regulator rings
Figure 1 showing the interactions among VRN1, VRN2, and VRN3 genes in a photoperiod-sensitive winter wheat. Arrows show induction, “T” show repression.
图 2 多年生黑麦草开花调控示意图
方框内容显示通路名称,箭头表示促进作用,“T”形表示抑制作用。春化途径中的基因以蓝色显示,光周期/生物钟途径中的基因以绿色显示,中央整合基因以红色显示。在花序中具有推定作用的MADS盒基因显示在左下,而来自自主途径的同源基因显示在右下。
Figure 2. Perennial ryegrass flowering control schematic diagram
Pathway names are placed in square boxes. Arrows show induction, “T” show repression. Genes in the vernalisation pathway are shown in blue, genes in photoperiod/circadian clock pathways in green, while central integrator genes are shown in red. MADS-box genes with putative role in floral identity are shown at the bottom left, while gene homologues from the autonomous pathway are shown at the bottom right.
图 3 鸭茅开花调控示意图
热图显示候选基因在不同阶段的相对表达,y轴代表已确定的候选基因,而x轴显示不同阶段。BV,春化前;V,春化期;AV,春化后;VG,营养生长期;BH,抽穗前;H,抽穗期。从BV到V定义为阶段1(P1),V到AV定义为第1阶段(P2),AV到VG定义为第2阶段(P3),VG到BH定义为第3阶段4(P4), BH到H定义为第5阶段(P5)。
Figure 3. Dactylis glomerata flowering control schematic diagram
The heat map shows relative expression of candidate genes at different stages. The y-axis represents identified candidate genes and the x-axis shows the different stages. BV, before vernalization; V, vernalization; AV, after vernalization; VG, vegetative growth; BH, before heading; H, heading. From BV to V is defined as phase 1 (P1), V to AV is defined as phase 2 (P2), AV to VG is defined as phase 3 (P3), VG to BH is defined as phase 4 (P4), and BH to H is defined as phase 5 (P5).
表 1 多年生黑麦草开花途径参与基因
Table 1 Genes involved in flowering pathway of Perennial ryegrass
基因名 Gene name 基因编号 GeneBank accession 同源基因 Homologous gene 通路/功能 Pathways/Function LpTFL1 AF316419 HvTFL1 抑制 Repressor LpSOC1 AtSOC1 中央整合子 Central integrator LpFT3 DQ309592 AtFT、OsHd3a、OsFTL2、HvFT、TaFT1 LpTSF AtFT LpSFT HvFT4 LpPRR37 OsPRR37 光周期 Photoperiod LpPRR73 OsPRR73 LpGI DQ534010 AtGI、HvGI、TaGI、OsGI LpPHYA AtPHYA LpPHYB AtPHYB LpPHYC PHYC LpCRY1 AtCRY1 LpCRY2 AtCRY2 LpTOC1 TOC1 LpCO AY600919 AtCO,OsHd1、HvCO1、TaHd1-1 (LpHD1) AM489608 Lpck2a-2 AB213317 Wheat tck2a 生物钟 Circadian clock Lpck2a-1 AB213316 OsCK2a LpCOL1 DQ145928 AtCO、OsHd1 LpLHY LHY LpLIR1 DQ206624 rice LIR LpGA20ox1 DQ071620 赤霉素 Gibberellin LpFLD FLD 自主促进 Autonomous LpFCA AY654582 AtFCA LpFY AtFY LpVrn2-2 DQ202716 TmVrn2 春化 Vernalization LpOX1 DQ145927 2OG-Fe(II) oxygenase LpJMJC JUMONJI-like LpVRN1 AY198326 TmVrn1、OsAP1、AtAP1、LtMADS1 (LpMADS1) GQ258851 LpVRN5 VRN5 LpMADS2 AY198327 AtAP1、LtMADS2 LpMADS3 AY198328 AtAP1、barley BM3 LpMADS4 AY198329 TaMADS12 花发育基因 Flower development LpMADS5 AY198330 barley BM9 LpMADS6 AY198331 OsMADS5 花发育基因 Flower development LpMADS7 AY198332 OsE31254 LpMADS8 AY198333 OsMADS24 LpMADS9 AY198334 Barley BM7 LpMADS10 DQ110009 OsMADS22、VRT2、SVP/AGL24-like 春化分生组织特性基因
Vernalization meristem identityLpMADS16 DQ110011 SVP-like 表 2 牧草开花基因
Table 2 Forage grass flowering gene
草种
Species开花基因
Flowering gene基因功能
Gene function参考文献
Reference多年生黑麦草
Lolium perenneLpFT3 由生物钟控制表达,与日长、春化时间相关,过度表达促进开花
Controlled by the circadian clock, related to the day length and the vernalisation time, over expression promotes flowering.[51] LpCO 长时间诱导,编码的蛋白促进开花
It is induced by long days and the encoded protein promotes flowering.[50] LpTFL1 与 FT 基因功能相反,参与开花抑制
In contrast to FT, it is involved in repression of flowering.[50] LpVRN1 春化诱导,表达模式与开花时间变化密切相关
It is induced by vernalisation and the expression pattern is correlated with variation in flowering time.[51] LpGI 与拟南芥 GI 同源,参与光周期开花时间控制
It is orthologous to Arabidopsis GI and involved in photoperiodic flowering time control.[17] LpHd1 水稻 OsHd1 的直系同源基因,影响开花时间
An ortholog of rice OsHd1 in affecting time of flowering.[49] 高羊茅
Festuca arundinaceaFaFT2 受光照调控,过表达促进开花
Controlled by light, over expression promotes flowering.[57] FaVRN1 开花诱导基因
Flowering inducing gene.[58] 草甸羊茅
Festuca pratensisFpVRN1 诱导 FT1 基因,在春季促进生殖器官的发育
FT1 gene expression is induced to promote development of reproduction organs in spring.[59] FpCO 直接结合 FT 基因的启动子以激活表达
Directly bind to the promoter of the FTgene to activate expression.FpFT1 促进开花
Promote floweringFpCK2α 在短日照条件下通过间接相互作用,增强 CO-like 蛋白质的功能
Enhances function of the CO-like protein through indirect interaction under short-day.扁穗冰草
Agropyron cristatumACOL CO 候选基因,在叶片中特异性表达
It is a CONSTANS (CO) candidate gene and showed specific expression in leaves.[60] 二穗短柄草
Brachypodium distachyonEZL1 拟南芥 CURLY LEAF 1 的直系同源基因,影响开花时间
An ortholog of Arabidopsis CURLY LEAF 1 in affecting flowering time.[61] 紫花苜蓿
Medicago sativaSPL13 调控开花时间和营养生长
It is regulating flowering time and vegetative growth.[62] miR156 过表达导致生物量产量加倍,开花时间延迟,纤维素含量增加和木质素含量减少Over expression resulted in a twofold increase in biomass yield, delayed flowering time, enhanced cellulose content, and reduced lignin. [63] 蒺藜苜蓿
Medicago truncatulaSTENOFOLIA(STF) 参与植株叶片的扩展,增加叶绿素含量,促进开花
It is involved in leaf expansion, increased chlorophyll content and accelerated flowering.[64] 红三叶
Trifolium pratenseWAT1 降低结实能力,减少种子产量
Reduces firmness and seed yield.[65] NIP4-1 提高种子产量
Increases seed yield.ZFP4 与结实相关
Closely related to seed setting.ERF106 控制种子重量
Controls seed weight.Pt4 提高种子产量
Increases seed yield.地三叶
Trifolium subterraneumCO-like 和 WNK5-like 蛋白介导远红光下的开花启动
Floral initiation under FR-enriched light is mediated by CO-like and WNK5-like proteins.[66] 白羽扁豆
Lupinus albus显性基因 Ef1 和 Ef2 当(Ef1_,Ef2_)同时出现时,表型早开花;(ef1ef1, ef2ef2)同时缺失时,表型晚开花;如果只有一个基因(Ef1_,ef2ef2 or ef1ef1, Ef2_)存在时,表型为中间体
When both dominant genes were present (Ef1_, Ef2_) the phenotype was early flowering, when both dominant genes were absent (ef1ef1, ef2ef2) the phenotype was late flowering and if only one gene was present (Ef1_, ef2ef2 or ef1ef1, Ef2_), the phenotype was intermediate.[67] -
[1] WOODS D P, MCKEOWN M A, DONG Y, PRESTON J C, AMASINO R M. Evolution of VRN2/GhD7-like genes in vernalization-mediated repression of grass flowering. Plant Physiology, 2016, 170(4): 2124-2135. doi: 10.1104/pp.15.01279
[2] DENG W, CASAO M C, WANG P, SATO K, HAYES P M, FINNEGAN E J, TREVASKIS B. Direct links between the vernalization response and other key traits of cereal crops. Nature Communication, 2015, 6(6): 5882.
[3] VOGEL K P, BREJDA J J, WALTERS D T, BUXTON D R. Switchgrass biomass production in the midwest USA. Agronomy Journal, 2002, 94(3): 413-420. doi: 10.2134/agronj2002.0413
[4] MOURADOV A, CREMER F, COUPLAND G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell, 2002, 14(Suppl): S111-S130.
[5] BOSS P K, BASTOW R M, MYLNE J S, DEAN C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell, 2004, 16(Suppl): S18-S31.
[6] PENG F Y, HU Z, YANG R C. Bioinformatic prediction of transcription factor binding sites at promoter regions of genes for photoperiod and vernalization responses in model and temperate cereal plants. BMC Genomics, 2016, 17(1): 573. doi: 10.1186/s12864-016-2916-7
[7] DORCA-FORNELL C, GREGIS V, GRANDI V, COUPLAND G, COLOMBO L, KATER M M. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant Journal for Cell & Molecular Biology, 2011, 67(6): 1006-1017.
[8] COCKRAM J, JONES H, LEIGH F J, O'SULLIVAN D, POWELL W, LAURIE D A, GREENLAND A J. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. Journal of Experimental Botany, 2007, 58(6): 1231-1244. doi: 10.1093/jxb/erm042
[9] HIGGINS J A, BAILEY P C, LAURIE D A. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One, 2010, 5(4): e10065. doi: 10.1371/journal.pone.0010065
[10] CHIRUTA C, FILIPOV F, CALIN M. Estimating the duration of daylight in a given time of the year depending on the latitude of the location. Research Journal of Agricultural Science, 2010, 42(3): 71-76.
[11] WADA M, SHIMAZAKI K, IINO M. Light Sensing in Plants. 2005. Berlin: Springer, 2005: 333-337.
[12] FERNÁNDEZ V, TAKAHASHI Y, LEGOURRIEREC J, COUPLAND G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. The Plant Journal, 2016, 86(5): 426-440.
[13] ONOUCHI H, IGEÑO M I, PÉRILLEUX C, GRAVES K, COUPLAND G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell, 2000, 12(6): 885-900. doi: 10.1105/tpc.12.6.885
[14] IZAWA T, MIHARA M, SUZUKI Y, GUPTA M, ITOH H, ATSUSHI J, NAGANO, MOTOYAMA R, SAWADA Y, YANO M, MASAMI YOKOTA HIRAI, MAKINO A, NAGAMURA Y. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell, 2011, 23(5): 1741-1755. doi: 10.1105/tpc.111.083238
[15] BENDIX C, MENDOZA J M, STANLEY D N, MEELEY R, HARMON F G. The circadian clock-associated gene gigantea1, affects maize developmental transitions. Plant Cell & Environment, 2013, 36(7): 1379-1390.
[16] MARTIN J, STORGAARD M, ANDERSEN C H, NIELSEN K K. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Molecular Biology, 2004, 56(2): 159-169. doi: 10.1007/s11103-004-2647-z
[17] GAGIC M, FAVILLE M, KARDAILSKY I, PUTTERILL J. Comparative genomics and functional characterisation of the GIGANTEA, gene from the temperate forage perennial ryegrass Lolium perenne. Plant Molecular Biology Reporter, 2015, 33(4): 1-9.
[18] HECHT V, LAURIE R E, WELLER J. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Frontiers in Plant Science, 2014, 5(17): 486.
[19] 王鹏, 张春庆, 陈化榜, 吴承来. 小麦冬性强弱评价体系的建立. 生态学报, 2012, 32(4): 1230-1240. WANG P, ZHANG C Q, CHEN H B, WU C L. The evaluation system of strength of winterness in wheat. Acta Ecologica Sinica, 2012, 32(4): 1230-1240.
[20] ZHANG X K, XIAO Y G, ZHANG Y, XIA X C, DUBCOVSKY J, HE Z H. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Science, 2008, 42(2): 1690-1694.
[21] 安艳荣. 二穗短柄草VILs基因对开花时间的调控机理研究. 青岛: 山东农业大学博士学位论文, 2015. AN Y R. Mechanism of regulation on flowering time by VIL genes in Brachypodium distachyon. PhD Thesis. Qingdao: Shandong Agricultural University, 2015.
[22] YOSHIDA T, NISHIDA H, ZHU J, NITCHER R, DISTELFELD A, AKASHI Y, KATO K, DUBCOVSKY J. Vrn-D4, is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theoretical & Applied Genetics, 2010, 120(3): 543-552.
[23] PIDAL B, YAN L, FU D, ZHANG, TRANQUILLI G, DUBCOVSKY J. The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. Journal of Heredity, 2009, 100(3): 355-364. doi: 10.1093/jhered/esp002
[24] DUBCOVSKY J, CHEN C, YAN L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Molecular Breeding, 2005, 15(4): 395-407. doi: 10.1007/s11032-005-0084-6
[25] YAN L, LOUKOIANOV A, BLECHL A, TRANQUILLI G, RAMAKRISHNA W, SANMIGUEL P, BENNETZEN J L, ECHENIQUE V, DUBCOVSKY J. The wheat VRN2 gene is a flowering repressor downregulated by vernalization. Science, 2004, 303(5664): 1640-1644. doi: 10.1126/science.1094305
[26] KIPPES N, ZHU J, CHEN A, VANZETTI L, LUKASZEWSKI A, NISHIDA H, KATO K, DVORAK J, DUBCOVSKY J. Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Molecular Genetics and Genomics, 2014, 289(1): 47-62. doi: 10.1007/s00438-013-0788-y
[27] MARQUARDT S, BOSS P K, HADFIELD J, DEAN C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. Journal of Experimental Botany, 2006, 57(13): 3379-3386. doi: 10.1093/jxb/erl073
[28] CHENG J Z, ZHOU Y P, LV T X, XIE C P, TIAN C E. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiology & Molecular Biology of Plants, 2017, 23(3): 1-9.
[29] SIMPSON G G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology, 2004, 7(5): 570-574. doi: 10.1016/j.pbi.2004.07.002
[30] EOM H, PARK S J, KIM M K, KIM H, KANG H, LEE I. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant Journal for Cell & Molecular Biology, 2017, 93(1): 79-91.
[31] YABUTA T, HAYASI T. Biochemical studies of 'bakanae' fungus of rice. Journal of the Imperial Agricultural Experimental Station Nisigahara Tokyo, 1940, 25(3): 365-400.
[32] 黄桃鹏, 李媚娟, 王睿, 李玲. 赤霉素生物合成及信号转导途径研究进展. 植物生理学报, 2015(8): 1241-1247. HUANG T P, LI M J, WANG R, LI L. Progress in study of gibberellins biosynthesis and signaling transduction pathway. Plant Physiology Journal, 2015(8): 1241-1247.
[33] BLAZQUEZ M A, GREEN R, NILSSON O, SUSSMAN M R, WEIGEL D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell, 1998, 10(5): 791-800. doi: 10.1105/tpc.10.5.791
[34] XUE L J, ZHANG J J, XUE H W. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PloS One, 2012, 7(2): e31081. doi: 10.1371/journal.pone.0031081
[35] 黄磊玉. 植物开花基因的研究进展. 农家科技, 2017(3): 257. HUANG L Y. Research progress of flowering genes in plants. Nongjia Keji, 2017(3): 257.
[36] 李昱, 罗志鹏, 赵淑清. 拟南芥开花时间调控的整合途径. 植物生理学报, 2007, 43(5): 799-804. LI Y, LUO Z P, ZHAO S Q. Integration pathway of flowering time control in Arabidopsis. Plant Physiology Journal, 2007, 43(5): 799-804.
[37] 袁敏, 邢朝斌, 葛伟娜, 王莉, 郭棣. 拟南芥开花诱导基因FT的蛋白表达及纯化. 基因组学与应用生物学, 2017, 36(8): 3053-3056. YUAN M, XING C B, GE W N, WANG L, GUO D. Protein expression and purification of flowering inducer gene FT in Arabidopsis thaliana. Genomics and Applied Biology, 2017, 36(8): 3053-3056.
[38] 兰树斌, 李建国. 植物LFY基因的研究进展. 基因组学与应用生物学, 2007, 26(s1): 132-137. LAN S B, LI J G. Advance of studies on LFY gene in plants. Genomics and Applied Biology, 2007, 26(s1): 132-137.
[39] 王利琳, 梁海曼, 庞基良, 朱睦元. 拟南芥LEAFY基因在花发育中的网络调控及其生物学功能. 遗传, 2004, 26(1): 137-142. doi: 10.3321/j.issn:0253-9772.2004.01.026 WANG L L, LIANG H M, PANG J L, ZHU M Y. Regulation network and biological roles of LEAFY in Arabidopsis thaliana in floral development. Hereditas, 2004, 26(1): 137-142. doi: 10.3321/j.issn:0253-9772.2004.01.026
[40] SAMACH A, ONOUCHI H, GOLD S E, DITTA G S, SCHWARZ-SOMMER Z, YANOFSKY M F, COUPLAND G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science, 2000, 288: 1613-1616. doi: 10.1126/science.288.5471.1613
[41] HEPWORTH S R, VALVERDE F, RAVENSCROFT D, MOURADOV A, COUPLAND G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo Journal, 2002, 21(16): 4327-4337. doi: 10.1093/emboj/cdf432
[42] SEARLE I, HE Y, TURCK F, VINCENT C, FORNARA F, KRÖBER S, AMASINO R A, COUPLAND G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development, 2006, 20(7): 898-912.
[43] SCHULTZ E A, HAUGHN G W. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell, 1991, 3(8): 771-781. doi: 10.1105/tpc.3.8.771
[44] LOHMANN J U, HONG R L, HOBE M, BUSCH M A, PARCY F, SIMON R, WEIGEL D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell, 2001, 105(6): 793-803. doi: 10.1016/S0092-8674(01)00384-1
[45] GUSTAFSON-BROWN C, SAVIDGE B, YANOFSKY M F. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell, 1994, 76(1): 131-143. doi: 10.1016/0092-8674(94)90178-3
[46] SHELDON C C, ROUSE D T, FINNEGAN E J, PEACOCK W J, DENNIS E S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C(FLC). Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(7): 3753-3758. doi: 10.1073/pnas.97.7.3753
[47] MICHAELS S D, AMASINO R M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 1999, 11(5): 949-956. doi: 10.1105/tpc.11.5.949
[48] PUTTERILL J, ROBSON F, LEE K, SIMON R, COUPLAND G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell, 1995, 80(6): 847-857. doi: 10.1016/0092-8674(95)90288-0
[49] SKØT L, SANDERSON R, THOMAS A, SKØT K, THOROGOOD D, LATYPOVA G, ASP T, ARMSTEAD I. Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiology, 2011, 155(2): 1013-1022. doi: 10.1104/pp.110.169870
[50] FIIL A, LENK I, PETERSEN K, JENSEN C S, NIELSEN K K, SCHEJBEL B, ANDERSEN J R, LÜBBERSTEDT T. Nucleotide diversity and linkage disequilibrium of nine genes with putative effects on flowering time in perennial ryegrass (Lolium perenne L.). Plant Science, 2011, 180(2): 228-237. doi: 10.1016/j.plantsci.2010.08.015
[51] WANG J, FORSTER J W. Flowering time regulation in perennial ryegrass. Euphytica, 2017, 213(5): 106. doi: 10.1007/s10681-017-1896-2
[52] XIE W G, ROBINS J G, BUSHMAN B S. A genetic linkage map of tetraploid orchardgrass (Dactylis glomerata L.) and quantitative trait loci for heading date. Genome, 2012, 55(5): 360-369. doi: 10.1139/g2012-026
[53] 谢文刚. 鸭茅分子遗传连锁图谱构建及开花基因定位. 成都: 四川农业大学博士学位论文, 2013. XIE W G. Genetic linkage map and flowering time gene mapping in orchardgrass (Dactylis glomerata L.). PhD Thesis. Chengdu: Sichuan Agricultural University, 2013.
[54] ZHAO X X, HUANG L K, ZHANG X Q, WANG J P, YAN D F, LI J, TANG L, LI X L, SHI T W. Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq. Scientific Reports, 2016, 6: 29345. doi: 10.1038/srep29345
[55] ZHAO X X, BUSHMAN B S, ZHANG X Q, ROBBINS M D, LARSON S R, ROBINS J G, THOMAS A. Association of candidate genes with heading date in a diverse Dactylis glomerata population. Plant Science, 2017, 265: 146-153.
[56] FENG G, HUANG L, LI J, WANG J, XU L, PAN L, ZHAO X, WANG X, HUANG T, ZHANG X. Comprehensive transcriptome analysis reveals distinct regulatory programs during vernalization and floral bud development of orchardgrass (Dactylis glomerata L.). BMC Plant Biology, 2017, 17(1): 216. doi: 10.1186/s12870-017-1170-8
[57] 陈锡, 赵德刚, 陈莹, 李小冬, 吴佳海, 王小利. 高羊茅FaFT2基因克隆及表达分析. 植物生理学报, 2017(8): 1523-1531. CHEN X, ZHAO D G, CHEN Y, LI X D, WU J H, WANG X L. Cloning and expression analysis of FaFT2 gene in tall fescue. Plant Physiology Journal, 2017(8): 1523-1531.
[58] 王小利, 陈伟, 李晚忱, 吴佳海, 刘晓霞, 杨义成. 高羊茅春化基因FaVRN1的克隆与分析. 核农学报, 2009, 23(5): 778-784. WANG X L, CHEN W, LI W C, WU J H, LIU X X, YANG Y C. Cloning and characterization of vernalizational gene FaVRN1 from tall fescue. Journal of Nuclear Agricultural Sciences, 2009, 23(5): 778-784.
[59] SHINOZUKA H, HAND M L, COGAN N O, SPANGENBERG G C, FORSTER J W. Nucleotide diversity of vernalization and flowering-time-related genes in a germplasm collection of meadow fescue (Festuca pratensis Huds.) Darbysh.). Ecology & Evolution, 2013, 3(13): 4415-4426.
[60] ZENG F, BILIGETU B, COULMAN B, SCHELLENBERG M P, FU Y B. RNA-Seq analysis of gene expression for floral development in crested wheatgrass (Agropyron cristatum L.). PLoS One, 2017, 12(5): e0177417. doi: 10.1371/journal.pone.0177417
[61] LOMAX A, WOODS D P, DONG Y, BOUCHÉ F, RONG Y, MAYER K S, ZHONG X, AMASINO R M. An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon. Plant Journal for Cell & Molecular Biology, 2018, 93(5): 871-882.
[62] GAO R, GRUBER M Y, AMYOT L, HANNOUFA A. SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Molecular Biology, 2018, 96(1/2): 1-15. doi: 10.1007/s11103-017-0689-2
[63] AUNG B, GRUBER M Y, HANNOUFA A. The MicroRNA156 system: A tool in plant biotechnology. Biocatalysis & Agricultural Biotechnology, 2015, 4(4): 432-442.
[64] LIU M, LEI L, MIAO F, POWERS C, ZHANG X, DENG J, TADEGE M, CARVER B F, YAN L. The STENOFOLIA gene from Medicago alters leaf width, flowering time and chlorophyll content in transgenic wheat. Plant Biotechnology Journal, 2018, 16(1): 186-196. doi: 10.1111/pbi.2018.16.issue-1
[65] KOVI M R, AMDAHL H, ALSHEIKH M, ROGNLI O A. De novo and reference transcriptome assembly of transcripts expressed during flowering provide insight into seed setting in tetraploid red clover. Scientific Reports, 2017, 7: 44383. doi: 10.1038/srep44383
[66] NAVARRO M P, RIBALTA F M, HURGOBIN B, CROSER J S, KAUR P. Gene networks underlying faster flowering induction in response to far-red light. BioRxiv, 2017.
[67] ADHIKARI K, BUIRCHELL B, YAN G, SWEETINGHAM M. Two complementary dominant genes control flowering time in albus lupin(Lupinus albus L.). Plant Breeding, 2011, 130(4): 496-499. doi: 10.1111/pbr.2011.130.issue-4
-
期刊类型引用(1)
1. 甄军波,宋世佳,刘琳琳,刘迪,欧阳艳飞,迟吉娜. 金花葵开花前后差异基因及代谢物分析. 华北农学报. 2023(S1): 58-66 .  百度学术
百度学术
其他类型引用(1)



 下载:
下载: