非生物胁迫下植物衰老和热激蛋白响应
English
-
植物生长发育常常受到土壤高盐度、干旱和极端温度等各种不利非生物胁迫因素的影响[1]。研究表明,非生物胁迫引起植物体内许多生理、生化和分子反应,进而影响植物产量和品质[2]。此外,遭受非生物胁迫的植物一般更容易受到病原、昆虫、杂草等生物胁迫的侵袭,极大地降低植物的生产能力[3]。植株衰老是一种高度有序的过程,在植株发育后期由外部环境和内部遗传因素共同控制,是植物细胞程序化死亡(programmed cell death, PCD)的一种[4-5]。在植物生长过程中,不利的非生物胁迫因素往往会提前启动叶片的衰老导致植物早熟减产、品质下降[6]。植物为适应恶劣的环境会加速衰老过程(应激诱导衰老),与植物在最适条件下表现出的自然衰老特征一致,发生典型应激反应的生理和分子现象[7]。研究表明所有的应激反应几乎都可以诱导产生热激蛋白(heat shock proteins, HSPs),它的转录受热应激转录因子(heat shock factors, HSFs)的调控[8]。热激蛋白可以以分子伴侣的身份协助蛋白质进行正确的组装或折叠,以阻止蛋白质出现不可逆聚集等,进而维持细胞在各种不良发育条件下的稳态[9]。在调控上,热激蛋白参与靶蛋白的降解和蛋白跨膜运输过程等多种生物功能[10]。研究表明,热激蛋白在高温、干旱、过氧化及重金属等非生物逆境下均能大量表达[11]。HSPs作为分子伴侣在保护植物免受胁迫和细胞内稳态重建方面发挥着关键作用[12]。牧草和草坪草作为植物中的一大类,在畜牧业发展、园林绿化和环境保护中具有非常重要的作用。为此,对非生物胁迫下牧草和草坪草等植物的衰老变化和热激蛋白响应进行综述,旨在为进一步研究植物非生物胁迫耐受性提供参考。
1. 非生物胁迫加速植物衰老
加速植株衰老和叶片脱落是植物逃避胁迫的主要策略之一,植物通过减少冠层大小来应对环境压力[13]。衰老过程中,植物将营养物质从源组织循环到生殖器官,这一循环过程由调节生理、生化和分子机制的复杂网络控制[14]。
1.1 衰老与光合作用
叶绿素存在于所有可以进行光合作用的生物体内。大多数情况下,植物体内叶绿素含量的多少可以衡量其生命的旺盛程度,一般情况下植物叶片中叶绿素含量越多,说明植物生命越旺盛,反之亦然。叶片衰老最显著、最初步的现象就是叶片黄化,主要原因是叶片衰老时叶绿素的降解先于类胡萝卜素[15]。植物体内全部叶绿素均以叶绿素–蛋白质复合物的形式存在于类囊体膜上,伴随着植物叶片的衰老,叶绿素降解酶活性大大增强,越来越多的叶绿素–蛋白质复合物被破坏,从而导致植物叶绿素含量降低。Graham等[16]指出,衰老过程中植物叶绿素降解速度明显大于合成速度,同时出现RNA大量水解、蛋白质迅速丧失、叶片黄化等特征,一些和光合作用过程相关的基因调控能力也会明显衰退。Giles等[17]研究认为,衰老叶片的光合作用能力降低与叶绿体结构的紊乱有关,光合作用速率下降的重要原因是叶绿体结构被破坏。植物衰老也会导致根系吸水能力减弱,使得养分和水分转移能力降低而使植物不能维持正常的生命代谢,随着胁迫加深,光合作用能力也将进一步下降[18]。弱光会加速拟南芥(Arabidopsis thaliana)叶片的衰老过程[19]。干旱胁迫下,复活草(Sporobolus stapfianus)叶片中叶绿素、类胡萝卜素和脂质含量都有所下降,衰老加快。
1.2 衰老与蛋白质含量
蛋白质是生物体的重要组成成分,蛋白质降解是植物衰老的基本特征之一。Feller和Keist[20]指出,叶绿体蛋白质的降解发生在衰老早期阶段,且叶绿素降解的过程与叶绿体蛋白质降解的途径部分相关。随着植物衰老,蛋白质合成速度下降,降解速度加快,从而导致体内蛋白质含量降低。Ray和Choudhuri等[21]研究指出,伴随着叶片衰老加剧,蛋白水解酶活力逐渐增加,使体内可溶性蛋白降解加快。Wilson等[22]研究认为,白三叶(Trifolium repens)叶片衰老过程中叶绿体的超微结构会发生显著变化,且在植物衰老后期叶绿体蛋白丰度会出现明显下降。Matile和Winkenbach等[23]研究认为,在正常条件下牵牛花(Ipomoea purpurea)体内的蛋白水解酶和底物各自分布在不同的区间,但当进入衰老状态时,蛋白水解酶会逐渐释放到细胞质中,从而加快蛋白质降解。一旦植物叶片进入衰老进程,蛋白质合成速度减缓,蛋白质水解酶活性上升,且蛋白质水解酶与底物在细胞空间内出现相互接近的现象,这三者共同影响了植物体内蛋白质的含量[24]。宋松泉等[25]对水稻(Oryza sativa)叶片进行的离体试验表明,随着水稻离体叶片衰老程度的加深,叶片中内肽酶和氨肽酶的活性呈显著增加的趋势,这一现象促使蛋白水解作用加强,因而降低了叶片中可溶性蛋白质含量。也有研究认为,叶片衰老时体内泛素相关基因的表达上升会加快蛋白水解速度[26]。热胁迫诱导匍匐剪股颖(Agrostis stolonifera)叶片衰老加快主要是由于叶绿素酶(chlorophyllase, Chlase)和脱镁叶绿素酶(pheophytinase, PPH)活性及其基因表达水平提高从而导致叶绿素降解加快所致,而与叶绿素合成无关。进一步研究发现热胁迫下拟南芥pph突变体的衰老显著慢于野生型,这证实了PPH可能是调节热胁迫加速叶绿素降解的关键酶之一[27]。此外,参与ATP代谢、光捕获、光呼吸、光化学反应以及活性氧清除的膜蛋白在调控热胁迫下匍匐剪股颖叶片的衰老过程中发挥着重要作用[28]。
1.3 衰老与抗氧化系统
持续的干旱、极端温度、高盐及黑暗等非生物环境胁迫在诱导植物早衰过程中经常出现活性氧(reactive oxygen species, ROS)积累的现象,而且会诱导超氧化物歧化酶(superoxide dismutase, SOD)、过氧化物酶(peroxidase, POD)、抗坏血酸过氧化物酶(ascorbate peroxidase, APX)以及过氧化氢酶(catalase, CAT)等抗氧化酶的活性发生变化[29]。非生物胁迫导致植物体内清除ROS的能力失衡,ROS过量积累会对细胞造成毒害作用,导致膜脂产生过氧化现象,致使维持细胞区域化的膜系统破损甚至瓦解,进一步加剧植物衰老速度。H2O2的过量生产加速了黑小麦(×Triticosecale wittmack)复水过程中的衰老,导致产量降低[30]。但植物在响应非生物胁迫过程中诱发产生的H2O2等ROS也可以作为信号分子激活衰老响应信号转导通路,并诱导抗氧化保护酶(APX、CAT、POD和SOD)基因的增强表达,提高胁迫下植物抗氧化清除能力[31]。过表达ROS清除酶能有效降低植物的氧化应激,增强植物耐受性,并延缓衰老[32]。Lee等[33]发现,干旱响应的NAC转录因子NTL4过表达时植株对干旱敏感,随着叶片中活性氧的积累,植物衰老的速度加快,然而NTL4缺失的突变体表现出叶片衰老延迟的现象,进而提高植物抗旱性。外源褪黑素(melatonin)通过激活多年生黑麦草(Lolium perenne)叶片中超氧化物歧化酶–过氧化氢酶途径及下调叶绿素降解来抑制暗诱导的叶片衰老[34]。
1.4 衰老与植物激素
植物激素在调节植物衰老过程中发挥着重要作用。一般来说,脱落酸(abscisic acid, ABA)、乙烯(ethylene, ETH)、茉莉酸(jasmonic acid, JA)和水杨酸(salicylic acid, SA)可以促进叶片衰老,而细胞分裂素(cytokinin, CTK)、生长素(auxin, IAA)和赤霉素(gibberellic acid)可以抑制叶片衰老[35]。
ABA是诱导植物衰老的一种重要内源激素,当植物衰老时体内的ABA合成代谢会被诱导,ABA含量大量增加。Zhao等[36]研究发现,ABA会通过激活蔗糖非发酵相关蛋白激酶(sucrose nonfermenting 1-related protein kinase, SnRK2s)促进叶片的衰老。Manoharan等[37]研究表明,由于添加了外源ABA,培养物在4周内达到最终衰老。衰老的标志是叶绿素的损失、蛋白质含量的下降和脂质组成发生变化。在干旱导致的拟南芥叶片衰老过程中出现ABA含量升高而CTK合成降低的现象[38]。He等[39]对早衰水稻新突变体(premature senescence leaf 85, psl85)的研究发现,psl85参与了ABA诱导叶片的衰老过程。外施ABA和乙烯释放剂均可高度诱导多年生黑麦草叶片中叶绿素分解代谢基因LpNYC1的表达并加速衰老,而乙烯生物合成抑制剂氨氧乙基乙烯基甘氨酸(aminoxyethyl vinyl glycine, AVG)抑制LpNYC1的表达[40]。多年生黑麦草中衰老正相关基因LpPPH介导的叶绿素(chlorophyll, Chl)降解可被ABA和乙烯正向调控,而被CTK负向调控,LpPPH可能是ABA和CTK信号通路中的直接下游靶基因[41]。研究证实,缺失ETH合成相关酶基因的拟南芥突变体叶片衰老速率显著降低[42]。另外,通过采用反义RNA技术抑制番茄(Lycopersicon esculentum)中的ACC氧化酶(1-aminocyclopropane-1-carboxylate oxidase)(参与ETH合成)活性,也可以通过降低ETH的合成延缓植物衰老的速度[43]。随着乙烯含量的增加,紫花苜蓿(Medicago sativa)叶片膜脂过氧化指数升高,过氧化伤害加大[44]。ETH调节PCD过程参与根皮质衰老(root cortical senescence, RCS)的过程[45]。
异戊烯基转移酶(isopentaltransferase, IPT)是限制催化CTK合成的第一个限速酶,IPT过表达的转基因植株表现出发育和形态上的改变并延缓叶片衰老[46]。CTK通过促进7-羟甲基叶绿素(7-hydroxymethyl chlorophyll, HmChl)积累和维持Chl a/b的比值延缓衰老,同时CTK处理还能维持较高的PSⅡ相关基因的转录水平,使水稻持绿性增强[47]。外源CTK通过减缓盐胁迫对多年生黑麦草的氧化和离子胁迫,从而有效降低衰老[48]。硅能促进CTK的生物合成,进而延缓拟南芥和高粱(Sorghum bicolor)的衰老[49]。外源CTK和ETH抑制剂处理均能显著抑制匍匐剪股颖叶片衰老,表现为热胁迫下草坪质量和叶绿素含量降低减缓[50]。SA会促进植物中叶绿素衰老相关基因(senescence-associated gene, SAGs)的表达,而SA合成途径关键基因(npr1和pad4)缺失的突变体中SAGs表达降低,衰老明显延缓[51]。Cheng等[52]通过全基因组研究发现,GA3处理能延缓黑暗诱导的结缕草(Zoysia japonica)幼苗叶片衰老,这与Met、SAM、V-ATPase、Cry (隐花色素基因)和An (白喉素合成酶基因)基因的差异表达有关。Zhang等[53]发现,SA和CTK可以提高热胁迫下多年生黑麦草叶绿素含量和光化学效率,同时降低电解质渗透率,增强多年生黑麦草的耐热性。近年来褪黑素在牧草和草坪草抗逆衰老中的研究较多,褪黑素可以抑制热诱导的多年生黑麦草叶片衰老,表现为衰老相关基因(LpSAG12.1和Lph36)转录水平的降低[54]。匍匐剪股颖中褪黑素减轻干旱胁迫引起的叶片衰老,这与叶绿素分解代谢基因的下调以及CTK合成基因和信号通路的协同作用有关[55]。热胁迫下外源褪黑素通过增加内源褪黑素和CTK含量,同时降低ABA含量,从而抑制多年生黑麦草叶片衰老[54]。
1.5 衰老与其他物质
植物衰老时,体内RNA和DNA含量都有所下降,但RNA下降速度高于DNA,其中rRNA含量的减少最为显著。在植物中,线粒体与叶绿体中的rRNA对衰老最为敏感,最先减少,而细胞质中的tRNA最后减少。Green[56]认为,rRNA含量明显下降与植物在衰老期间细胞内RNA酶活性增强密切相关。随着衰老程度的加深,一些mRNA的数量减少甚至消失,然而另外一些mRNA的数量则会增加。这种现象表明叶片在衰老过程中一部分基因表达水平会受到抑制,甚至完全不表达,而另一些基因在衰老时又会被激活,这与基因功能息息相关。衰老时细胞膜的功能完整性和结构都会趋于衰退,膜脂降解速度加快,这与脂代谢相关酶(如脂氧化酶、裂解酰基水解酶、磷脂酸磷酸酶以及磷脂酶D)基因在衰老时的促进表达有关。成熟叶片过氧化物酶体中具有催化脂肪酸β-氧化的酶类,当衰老造成碳水化合物供应不足时,这一催化反应途径就至关重要[57]。对拟南芥和大麦(Hordeum vulgare)的研究表明,衰老叶片中与脂类降解相关的乙醛酸循环代谢中的关键酶(如异柠檬酸裂解酶及苹果酸合成酶)活性和含量升高[58]。Doorn等[59]在研究鸢尾(Iris tectorum)花的衰老时发现,磷脂酶D的活性在开花当天大量增加,在肉眼可见衰老前磷脂就已经开始降解。伴随着叶片的衰老,细胞内含物大量水解,还原糖、氨基酸含量上升[60]。干旱胁迫下,复活草光呼吸通路中酶活性、氨基酸和糖类含量均有一定程度的下降[61]。外源施用柠檬酸可能有利于维持膜的稳定性、根的活性、抗氧化反应和HSP基因的激活,缓解高温对高羊茅(Lolium arundinaceum)生长和生理代谢的损伤[62]。近年有研究表明生物钟在拟南芥衰老过程中也起到关键作用[63]。
1.6 植物衰老的分子调控
在植物衰老的不同阶段,有一类基因会被诱导上调表达,这些基因被称为SAGs,SAGs只在衰老过程开始后才表达,不参与衰老过程的诱导[64-65]。胁迫诱导衰老的调控被认为是一种可行的植物抵抗胁迫产生的策略,一些基因/通路被证明与调控这一过程有关[66]。各种调节SAGs的转录因子在不同衰老时间点被激活,这些转录因子大多可分为NAC和WRKY两种。Uauy等[67]研究表明,通过RNA干扰降低多个NAM (NAC转录因子)同源物的RNA水平,可延缓衰老3周以上,小麦(Triticum aestivum)籽粒蛋白质、锌、铁含量提高30%以上。Ma等[68]研究表明,SlNAP2 (一种NAC转录因子)在番茄叶片衰老和果实产量控制中起着重要作用。GhWRKY42可被叶片衰老和各种胁迫诱导,然而GhWRKY42基因如何调控叶片衰老还需进一步的研究和阐明[69]。拟南芥中与衰老相关的转录因子有NAC、WRKY、MYB、C2H2锌指蛋白、bZIP和AP2/EREBP家族。转录因子OsWRKY5通过调节衰老相关基因NAC和ABA生物合成途径促进水稻叶片衰老[70]。水稻MYB转录因子OsMYB102通过下调ABA的生物合成和下游信号通路参与了叶片衰老的调控[71]。拟南芥中AtMYB44转录因子的调控功能可能是高度保守的,他们在叶片衰老和非生物胁迫反应中起着重要的作用[72]。拟南芥、大豆(Glycine max)和水稻中与衰老相关的受体激酶(senescence-associated receptor-like kinase, SARK)家族成员被认为是叶片衰老的积极调节因子。Li等[73]研究表明苔藓(Physcomitrella patens)中衰老相关受体激酶(senescence-associated receptor-like kinase, PpSARK)在盐胁迫反应中起正向调节作用,而在衰老过程中可能起负向调节作用。S40基因家族在大麦、拟南芥和水稻衰老过程中起着重要作用[74-75]。过表达玉米(Zea mays)转录因子ZmVQ52可加速拟南芥叶片衰老,相比于野生型表现出更低的叶绿素含量和更快的衰老速度[76]。棉花(Gossypium hirsutum)中衰老诱导的ZmSUT1的表达延缓了叶片的衰老,而种皮中特异性表达ZmSUT1能提高棉花纤维产量和籽粒产量[77]。Sekhon等[78]通过系统的基因组和转录组分析发现,玉米衰老过程中涉及蛋白质水解(半胱氨酸蛋白酶)、糖信号、糖转运、糖的卸载到碳汇和细胞壁多糖的合成等多种过程。其中大多数基因都是新的候选基因,尽管根据已发表的文献发现他们与衰老之间存在逻辑联系,但还没有经过验证,其中一半的候选基因在拟南芥中被认为是SAGs。而迄今为止,在草类中只有NAC家族的转录因子直接通过遗传研究证实参与衰老的调节[79]。过表达持绿基因LpSGR的多年生黑麦草叶片中Chl含量增加,光化学效率提高,叶片衰老减缓[80]。LpNYC1、LpNOL和LpPPH等叶绿素分解基因的转录水平与热诱导的叶片衰老呈负相关关系,下调这些基因的表达可抑制热诱导的叶片衰老和氧化损伤,使多年生黑麦草耐热性增强[53]。
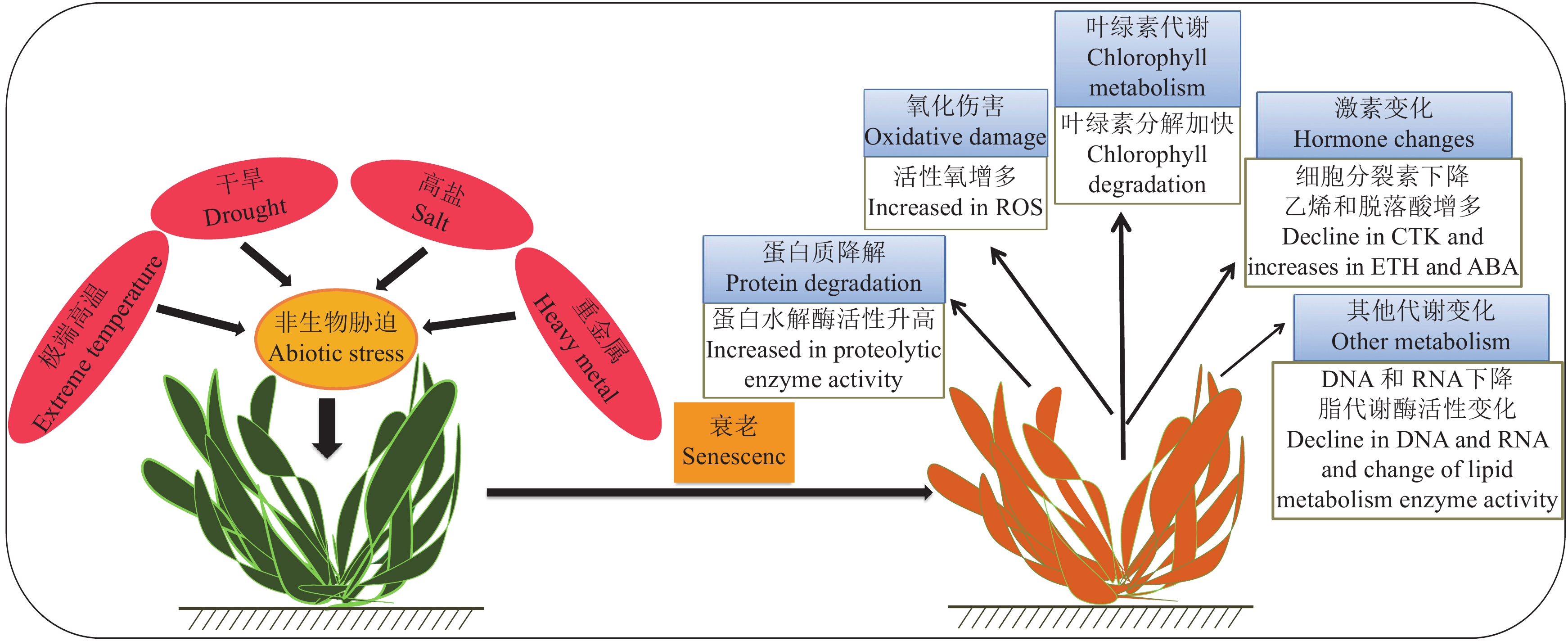
衰老是一个复杂的、高度调节的过程,发生的化学变化、结构变化和代谢变化涉及到数千个基因的表达调控,这些基因在衰老过程中下降或上升。随着叶片衰老过程的进行,基因表达的上调和下调网络相互作用明显减少[81]。每一个促进衰老的因子都会上调一组与衰老相关的基因,这些基因依次参与感知、信号转导通路和最终反应,所有基因都受到复杂的调控串扰,图1总结了非生物胁迫诱导的植物衰老响应。
2. 非生物胁迫下热激蛋白响应
2.1 热激蛋白分类及其主要功能
HSPs存在于所有的生物体中,是一种与应激诱导的其他蛋白变性相反的分子伴侣蛋白,在生物和非生物胁迫耐受中起着关键作用[82]。除可以帮助新合成的蛋白质有效折叠外,还能使已经存在的蛋白质保持稳定构象,进一步防止他们在压力条件下聚集[83]。不同细胞类型的HSPs在生物体之间甚至在生物体内部的数量均存在差异。Rao等[84]根据HSPs分子量大小将其分为6大家族,分别为HSP100、HSP90 (83~90 kD)、HSP70 (66~78 kD)、HSP60 (60 kD)、小热激蛋白(small heat shock proteins, sHSPs) (15~42 kD)和泛素(8.5 kD) (表1)。
2.1.1 HSP100
酪蛋白溶解蛋白酶B (casein lytic proteinase B, Clp B)是分子量最大的HSPs (约100 kD),属于HSP100家族,因此HSP100又称为Clp B[85]。HSP100可分为质体型HSP100和胞质型HSP100。其中胞质型HSP100只有在热激条件下才会产生,在正常条件下以非必需蛋白的形式存在。HSP100最先溶解异常蛋白以维护蛋白质的稳态,溶解异常蛋白后交给HSP70系统进行重新折叠和加工,最终在HSP60作用下再次形成具有正常功能的蛋白[86]。在整个植物生长过程中,HSP100既受生长发育调控,同时也受热激、寒冷、干旱和高盐等胁迫的诱导[87-88]。Grover等[89]报道了转HSP100基因的烟草、水稻和拟南芥均表现出耐热性增加的现象,表明HSP100对提高植物耐热能力具有促进作用。
表 1 热激蛋白分类及功能Table 1. Classification and functions of heat shock proteins热激蛋白分类
Classification of heat shock protein主要功能
Main functionHSP100 溶解变性蛋白
Dissolved denatured proteinHSP90 (83~90 kD) 促进蛋白质成熟、折叠和组装
Promotes ripening, folding assemblyHSP70 (66~78 kD) 防止聚集,协助重新折叠,蛋白质导入和易位,信号转导和转录激活
Prevents aggregation, assists in refolding, protein introduction and
translocation, signal transduction, and transcriptional activationHSP60 (60 kD) 蛋白质折叠与辅助折叠
Folding and auxiliary foldingsHSPs (15~42 kD) 防止聚合和稳定异常蛋白
Prevents polymerization and stabilizes abnormal proteins泛素 Ubiquitin (8.5 kD) 标记需要分解的蛋白,参与蛋白质降解
To label proteins that need to be broken down and participates in protein degradation2.1.2 HSP90
HSP90是一组高度保守的分子伴侣,是蛋白质稳态网络中的一个多功能枢纽,也是所有已检测的真核细胞中细胞生存所必需的分子伴侣。一般来说,HSP90在蛋白质折叠的后期起作用,调控多种蛋白质的折叠、成熟和稳定性或稳定多亚基蛋白复合物成分,促进其组装[90-92]。HSP90具有较高的识别特异性,这得益于其复杂的协同伴侣系统。HSP90及其协同伴侣参与细胞对非生物胁迫的反应和分化,对这些功能的调节至关重要[93]。Xu等[94]用拟南芥5个典型基因(GmHsp90A2、GmHsp90A4、GmHsp90B1、GmHsp90C1.1、GmHsp90C2.1)的过表达验证了GmHsp90基因能降低植株的非生物胁迫损伤,提高植株抗逆能力。Zhang等[95]研究结果表明,BdHsp90参与二穗短柄草(Brachypodium distachyon)非生物胁迫响应。HSP90在真菌和动物体内的结构、进化和功能已被详细研究,而植物中关于HSP90的研究仍然比较有限。Virdi等[96]发现高粱中HSP85可能是HSP90家族的一员。
2.1.3 HSP70
HSP70又称为热激同源蛋白(heat shock cognates, HSC),是HSPs家族中最保守的一类,同时也是真核细胞中最丰富的HSPs之一。HSP70的主要功能是参与新生蛋白的折叠,也可以和部分蛋白疏水区结合以阻止蛋白聚合,参与蛋白的跨膜运输和靶蛋白的降解过程。HSP70可以由所有导致蛋白质变性的胁迫引起[97]。HSP70在受到多种非生物胁迫时发挥重要作用[98]。小麦叶片中分离的HSP70基因(TaHSC70)在高温胁迫和条状锈菌感染引起的防御反应中发挥作用,并通过依赖于茉莉酸的信号转导途径发挥作用[99]。水稻中mtHsp70可能通过维持线粒体膜电位和防止活性氧扩增来抑制线粒体原生质体程序性死亡[100]。转基因上调HSP70已被证实可以增强植物对高温[101-102]、干旱[103]和盐度[104]的耐受性。
2.1.4 HSP60
HSP60家族包括真核生物中HSP60和原核生物中蛋白(groel, Gro EL),他们具有高度的保守性。HSP60可以和很多不同的蛋白结合,进而防止蛋白在转录之后与折叠前出现异常聚集情况[105]。Haq等[106]研究发现,辣椒(Capsicum annuum)中CaHSP60-6基因的敲除增加了热胁迫敏感性,表现为较高的相对电解质泄漏、脂质过氧化、活性氧种类的积累以及较低的叶绿素含量和抗氧化酶活性。这些结果表明,HSP60可能在辣椒热防御和其他非生物胁迫中起着积极的调节作用。HSP60在多种应激中作用[107]。Tominaga等[108]从孔石莼(Ulva lactuca)中分离出Up HSP60基因,研究结果表明其基因的表达水平受温度、日照以及重金属的影响,推测出HSP60可能参与温度、光照和重金属胁迫应激反应。
2.1.5 sHSPs和泛素
sHSP是在植物中普遍存在的一种较为复杂的小分子热激蛋白,通常形成分子量为200~800 kD的寡聚物。在细胞伴侣蛋白网络中,sHSPs是蛋白发生错误折叠的第一道防线,又被称为细胞卫士[109-110]。sHSPs的主要功能是通过非极性方式与异常蛋白结合,从而阻止异常蛋白的相互聚集和异常蛋白溶解,但sHSPs本身并不参与错误蛋白的再折叠过程,只是首先通过分子伴侣与之结合,再经过HSP70/HSP100完成整个加工过程[111]。sHSPs不仅会在植物生长的不同生长期产生[112],还会在受到干旱、高温、盐、寒冷等逆境胁迫时诱导表达[113]。Sato和Yokoya[114]研究表明,水稻中过表达sHsp17.7基因可显著提高转基因水稻幼苗的耐旱性。OsMSR3(sHSPs)通过增强抗氧化防御机制和ABA响应的基因表达提高拟南芥对铜胁迫的耐受性[115]。Redddy等[116]结果表明,御谷(Pennisetum glaucum)叶片中CI-sHsp (胞质类I)基因在不同的非生物胁迫条件下有不同的表达。匍匐剪股颖中AsHSP17 (sHSPs)可能通过调节光合作用和ABA依赖或独立的信号通路,作为蛋白伴侣负调控植物对非生物胁迫的响应[117]。
泛素是一种高度保守的蛋白质,含有76种氨基酸,泛素水平的增加是由于热胁迫加强,降解受损蛋白质的需求增加。Zhang等[118]研究表明,非胁迫状态下的豆叶中泛素的转录水平较低,而金属(如Hg、Cd、As、Zn、Cu)、温度升高、病毒感染、水杨酸等均强烈刺激了基因的表达,这表明泛素蛋白水解系统可能在抗各种环境胁迫中发挥重要作用。Belknap和Garbarino[119]研究表明,泛素途径参与植物衰老和应激反应,泛素在衰老过程中的作用可能是促进蛋白质体降解进行氮循环。Irene等[120]研究表明,E3的泛素连接酶蛋白作为泛素化级联的主要成分,赋予底物识别的特异性。拟南芥E3连接酶参与ABA通路响应干旱和盐胁迫[121]。
2.2 热激转录因子调节热激蛋白应答非生物胁迫
HSFs可作为热激基因和HSPs以及其他分子伴侣表达的调控因子[122]。应激条件下HSPs/分子伴侣受多种HSFs的控制[123-124]。Shim等[125]研究表明,小麦和水稻的HsfA4a通过上调植物体内MT基因的表达来增强对Cd的耐受性。所有真核生物中都存在HSFs,对植物中HSFs序列和表达模式的比较发现其在抗胁迫和调控发育方面具有重叠作用[126-127]。HSPs序列上游启动子存在热应答元件(heat shock element, HSE),HSFs可以特异性识别HSPs基因上游的HSE进而调控HSPs的表达。一般情况下,HSFs是以单体形式与阻遏蛋白结合,但不能和HSE结合,应激情况导致HSFs与阻遏蛋白分离,同时在HSE附近形成三聚体进而与HSE进行特异性结合,促进HSPs基因转录的起始。研究发现,非胁迫条件下HSFs可以与HSP70相互作用而保持HSFs的非活性状态,当受到逆境胁迫时,异常蛋白会与HSP70竞争结合HSFs,解除抑制的HSFs可以形成具有DNA结合活性的三聚体,通过与HSE结合从而启动HSPs基因的转录[128](图2)。
植物中HSPs分子伴侣与其他应激反应机制可能存在交叉作用。例如,HSPs/分子伴侣可以在应激信号转导和基因激活中发挥作用[129],也可以调节细胞氧化还原状态[130]。他们还与其他应激反应机制相互作用,如渗透物和抗氧化剂[131-132]。不同种类的HSPs/分子伴侣在保护蛋白质免受胁迫方面发挥着互补和重叠的作用。在维持细胞内稳态过程中,一些HSPs家族成员(HSP70和sHSPs)可以稳定蛋白质的构象,防止聚集,进而使非原生蛋白保持在一种能够使HSPs (如HSP60、HSP70及HSP90)进行后续折叠的状态。当变性或错误折叠的蛋白质形成聚合体时,他们可以被HSP100溶解后再重新折叠,或被蛋白酶降解。一些HSPs(如HSP70和HSP90)伴随信号转导和转录激活,从而导致HSPs其他成员的合成(如HSP70和HSP90),主要受HSFs和其他应激反应蛋白(如抗氧化物酶)调控[86]。Wang等[133]通过RNA-Seq分析高羊茅和多年生黑麦草HSFs对温度胁迫的响应,为今后从基因功能及转基因方面研究草类植物和其他作物的抗逆性奠定了基础。植物获得耐受性往往是各种生理生化反应的结果,这些机制可协同作用以防止细胞损伤与重建体内的平衡[134]。HSPs是一种抗逆蛋白,不同家族间相互作用共同构建一个蛋白网络来维护其他蛋白稳态。柳枝稷(Panicum virgatum)差异表达基因中有21个HSFs和22个HSPs在Cd处理后表达水平升高,在拟南芥中过表达1个HSP编码基因可以显著提高对其Cd的耐受性[135]。对HSP100、HSP90、HSP70和sHSPs的表达模式研究表明,中等氮(每14 d 7.5 kg·hm−2)水平促进了长时间热胁迫下HSP100、HSP90、HSP70和sHSPs的产生,有助于匍匐剪股颖更好地经受长期的热胁迫[136]。
3. 总结与展望
高温、盐胁迫和干旱等非生物胁迫下植物激活相应的细胞信号通路和细胞反应,如通过HSPs产生、抗氧化防御增强和细胞渗透调节溶质积累等途径来提高自身抗性。植物在响应非生物胁迫过程中最初的应激信号(温度、离子效应、渗透性、水分和膜流动性)触发下游信号传递和转录调控,激活应激反应机制,以重新建立稳态。但非生物胁迫常导致细胞内稳态发生不可逆变化,并破坏其功能和结构蛋白,细胞膜氧化损伤,导致细胞衰老甚至死亡。植物可通过调节代谢物、抗氧化防御、激素水平等延缓逆境对植物造成的衰老或通过加速衰老以适应环境。植物体内的HSPs作为植物逆境应激蛋白在非生物胁迫中起着重要的调节作用,可延缓植物衰老,并增强植物非生物胁迫抗性。关于植物衰老响应及热激蛋白通路的研究对了解植物适应性机制、提高植物抗逆性、培育抗逆作物及降低环境因素对植物的伤害等方面意义重大,也将是植物领域研究的重点和难点。未来可从激素互作、衰老调控关键基因克隆和遗传转化、筛选晚衰新种质等方面研究非生物胁迫下植物的衰老响应,为培育高产抗逆的牧草或持绿性好的草坪草寻找新的途径。
参考文献
[1] ZHU J K. Abiotic stress signaling and responses in plants. Cell, 2016, 167(2): 313-324. doi: 10.1016/j.cell.2016.08.029
[2] WANG W, VINOCUR B, ALTMAN A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta, 2003, 218(1): 1-14. doi: 10.1007/s00425-003-1105-5
[3] KHAN S A, LI M Z, WANG S M, YIN H J. Revisiting the role of plant transcription factors in the battle against abiotic stress. International Journal of Molecular Sciences, 2018, 19(6): 1634. doi: 10.3390/ijms19061634
[4] SMART C M. Gene expression during leaf senescence. New Phytologist, 2006, 126(3): 419-448.
[5] GAN S, AMASINO R M, AMASINO R M. Making sense of senescence: Molecular genetic regulation and manipulation of leaf senescence. Plant Physiology, 1997, 113(2): 313-319. doi: 10.1104/pp.113.2.313
[6] GAN S. Mitotic and postmitotic senescence in plants. Science of Aging Knowledge Environment, 2003, 2003(38): 7.
[7] NOODÉN L D, GUIAMÉT J J, JOHN I. Senescence mechanisms. Physiologia Plantarum, 2010, 101(4): 746-753.
[8] SCHARF K D, BERBERICH T, EBERSBERGER I, NOVER L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochimica Biophysica Acta, 2018, 1819(2): 104-119.
[9] REDDY P S, MALLIKARJUNA G, KAUL T, CHAKRADHAR T, MISHRE R N, SOPORY S K, REDDY M K. Molecular cloning and characterization of gene encoding for cytoplasmic Hsc70 from Pennisetum glaucum may play a protective role against abiotic stresses. Molecular Genetics and Genomics, 2010, 283(3): 243-254. doi: 10.1007/s00438-010-0518-7
[10] BOSTON R S, VIITANEN P V, VIERLING E. Molecular chaperones and protein folding in plants. Plant Molecular Biology, 1996, 32(1/2): 191-222. doi: 10.1007/BF00039383
[11] FU X, ZOU Z. Abiotic regulation: A common way for proteins to modulate their functions. Current Protein and Peptide Science, 2015, 16(3): 188-195. doi: 10.2174/1389203716666150224124429
[12] WATERS E R, LEE G J, VIERLING E. Evolution, structure and function of the small heat shock proteins in plants. Journal of Experimental Botany, 1996, 47(3): 325-338. doi: 10.1093/jxb/47.3.325
[13] KOOYERS, NICHOLAS J. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Science, 2015, 234: 155-162. doi: 10.1016/j.plantsci.2015.02.012
[14] MUNNÉ-BOSCH S, ALEGRE L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Functional Plant Biology, 2004, 31(3): 8808-8818.
[15] KRUPINSKA K. Fate and activities of plastids during leaf senescence. The Structure and Function of Plastids, 2007, 23: 433-449.
[16] GRAHAM I A, LEAVER C J, SMITH S M. Induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell, 1992, 4(3): 349-357. doi: 10.2307/3869545
[17] GILES K L, COHEN D, BEARDSELL M F. Effects of water stress on the ultrastructure of leaf cells of sorghum bicolor. Plant Physiology, 1976, 57(1): 11-14. doi: 10.1104/pp.57.1.11
[18] 杨树勋. 植物叶片衰老机理及在烟叶生产上的应用. 作物研究, 2018, 32(1): 90-96. YANG S X. Mechanisms of plant senescence and their application in tobacco leaf production. Crop Research, 2018, 32(1): 90-96.
[19] JANEČKOVÁ H, HUSIČKOVÁ A, FERRETTI U, PRČINA M, PILAROVÁ E, PLAČKOVÁ L, POSPÍŠIL P, DOLEŽAL K, ŠPUNDOVÁ M. The interplay between cytokinins and light during senescence in detached Arabidopsis leaves. Plant, Cell and Environment, 2018, 41(8): 1870-1885. doi: 10.1111/pce.13329
[20] FELLER U, KEIST M. Senescence and Nitrogen Metabolism in Annual Plants. Berlin: Springer Netherlands. 1986: 219-234.
[21] RAY S, CHOUDHURI M A. Flag leaf senescence in intact rice plant: Effects of hormones on the activities of “senescence-enzymes” during leaf age at the reproductive development. Biochemie Und Physiologie Der Pflanzen, 1980, 175(4): 346-353. doi: 10.1016/S0015-3796(80)80075-8
[22] WILSON K A, MCMANUS M T, GORDON M E, JORDAN T W. The proteomics of senescence in leaves of white clover, Trifolium repens (L.). Proteomics, 2002, 2(9): 1114-1122. doi: 10.1002/1615-9861(200209)2:9<1114::AID-PROT1114>3.0.CO;2-O
[23] MATILE P, WINKENBACH F. Function of lysosomes and lysosomal enzymes in the senescing corolla of the morning glory (Ipomoea purpurea). Journal of Experimental Botany, 1971, 22(4): 759-771. doi: 10.1093/jxb/22.4.759
[24] MARTIN C, THIMANN K V. Role of protein synthesis in the senescence of leaves: II. The influence of amino acids on senescence. Plant Physiology, 1972, 50(4): 432-437. doi: 10.1104/pp.50.4.432
[25] 宋松泉, 苏卫珍, 彭晓南. 杂交水稻离体叶片衰老与蛋白质代谢的关系. 逻辑学研究, 1995(1): 20-23. SONG S Q, SU W Z, PENG X N. Relationship between leaf senescence and protein metabolism in vitro of hybrid rice. Studies in Logic, 1995(1): 20-23.
[26] GLICKMAN M H, CIECHANOVER A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews, 2002, 82(2): 373-428. doi: 10.1152/physrev.00027.2001
[27] JESPERSEN D, ZHANG J, HUANG B. Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Science, 2016, 249: 1-12. doi: 10.1016/j.plantsci.2016.04.016
[28] JESPERSEN D, XU C, HUANG B C. Membrane proteins associated with heat‐induced leaf senescence in a cool‐season grass species. Crop Science, 2015, 55(2): 837-850. doi: 10.2135/cropsci2014.04.0335
[29] YAN C S, LI D Q, ZHANG J H. Plant leaf senescence and oxidative stress. Chinese Bulletin of Botany, 2005, 10(3): 515-534.
[30] HURA T, HURA K, OSTROWSKA A, GADZINOWSKA J, FIUST A. Water stress-induced flag leaf senescence may be accelerated by rehydration. Journal of Plant Physiology, 2019, 236: 109-116. doi: 10.1016/j.jplph.2019.01.013
[31] MITTLER G, SUZUKI N, CIFTCI-YILMAZ S, MITTLER R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell and Environment, 2010, 33(4): 453-467. doi: 10.1111/j.1365-3040.2009.02041.x
[32] TSENG M J, LIU C W, YIU J C. Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiology and Biochemistry, 2007, 45(10/11): 822-833. doi: 10.1016/j.plaphy.2007.07.011
[33] LEE S, SEO P J, LEE H J, PARK C M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant Journal, 2012, 70(5): 831-844. doi: 10.1111/j.1365-313X.2012.04932.x
[34] ZHANG J, LI H, XU B, LI J, HUANG B. Exogenous melatonin suppresses dark-induced leaf senescence by activating the superoxide dismutase-catalase antioxidant pathway and down-regulating chlorophyll degradation in excised leaves of perennial ryegrass (Lolium perenne L.). Frontiers in Plant Science, 2016, 7: 1500.
[35] JIBRAN R, HUNTER D, DIJKWEL P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Molecular Biology, 2013, 82(6): 547-561. doi: 10.1007/s11103-013-0043-2
[36] ZHAO Y, CHAN Z, GAO J, XING L, CAO M, YU C, HU Y, YOU J, SHI H, ZHU Y, GONG Y, MU Z, WANG H, DENG X, WANG P, BRESSAN R A, ZHU J K. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(7): 1949-1954. doi: 10.1073/pnas.1522840113
[37] MANOHARAN K, PRASAD R, GUHA-MUKHERJEE S. Senescence-related lipid changes in callus cultures of Datura innoxia. Phytochemistry, 1990, 29(8): 2529-2531. doi: 10.1016/0031-9422(90)85181-E
[38] DONG H, NIU Y, LI W, ZHANG D. Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. Journal of Experimental Botany, 2008, 59(6): 1295-1304. doi: 10.1093/jxb/ern035
[39] HE Y, ZHANG Z, LI L, TANG S, WU J L. Genetic and physio-biochemical characterization of a novel premature senescence leaf mutant in rice (Oryza sativa L.). International Journal of Molecular Sciences, 2018, 19(8): 2339. doi: 10.3390/ijms19082339
[40] XU B, LI H, YU G, ZHANG J, HUANG B. Characterization and transcriptional regulation of chlorophyll b reductase gene NON-YELLOW COLORING 1 associated with leaf senescence in perennial ryegrass (Lolium perenne L.). Environmental and Experimental Botany, 2018, 149(1): 43-50.
[41] JING Z, YU G, WEN W, MA X, HUANG B. Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. Journal of Experimental Botany, 2015, 67(3): 935-945.
[42] OH S A, PARK J H, LEE G I, PAEK K H, PARK S K, NAM H G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. The Plant Journal, 2003, 12(3): 527-535.
[43] JOHN I, DRAKE R, FARRELL A, COOPER W, LEE P, HORTON P, GRIERSON D. Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: Molecular and physiological analysis. The Plant Journal, 1995, 7: 483-490. doi: 10.1046/j.1365-313X.1995.7030483.x
[44] IRIGOYEN J J, EMERICH D W, SÁNCHEZ-DÍAZ M. Alfalfa leaf senescence induced by drought stress: Photosynthesis, hydrogen peroxide metabolism, lipid peroxidation and ethylene evolution. Physiologia Plantarum, 2006, 84(1): 67-72.
[45] SCHNEIDER H M, TOBIAS W, POSTMA J A, BROWN K M, LYNCH J P. Ethylene modulates root cortical senescence in barley. Annals of Botany, 2018, 122(1): 95-105. doi: 10.1093/aob/mcy059
[46] GAN S, AMASINO R M. Cytokinins in plant senescence: From spray and pray to clone and play. Bioessays, 1996, 18(7): 557-565. doi: 10.1002/bies.950180707
[47] TALLA S K, PANIGRAPHY M, KAPPARA S, NIROSHA P, NEELAMRAJU S, RAMANAN R. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. Journal of Experimental Botany, 2016, 67(6): 1839-1851. doi: 10.1093/jxb/erv575
[48] MA X, ZHANG J, HUANG B. Cytokinin-mitigation of salt-induced leaf senescence in perennial ryegrass involving the activation of antioxidant systems and ionic balance. Environmental and Experimental Botany, 2016, 125: 1-11. doi: 10.1016/j.envexpbot.2016.01.002
[49] MARKOVICH O, STEINER E, KOURIL Š, TARKOWSKI P, AHARONI A, ELBAUM R. Silicon promotes cytokinin biosynthesis and delays senescence in Arabidopsis and Sorghum. Plant, Cell and Environment, 2017, 40(7): 1189-1196. doi: 10.1111/pce.12913
[50] JESPERSEN D, YU J, HUANG B. Metabolite responses to exogenous application of nitrogen, cytokinin, and ethylene inhibitors in relation to heat-induced senescence in creeping bentgrass. PLoS One, 2015, 10(3): e0123744. doi: 10.1371/journal.pone.0123744
[51] GUO P, LI Z, HUANG P, LI B, FANG S, CHU J, GUO H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. The Plant Cell, 2017, 29(11): 2854-2870. doi: 10.1105/tpc.17.00438
[52] CHENG X, DAI X, ZENG H, LI Y, WEI T, HAN L. Gene expression involved in dark-induced leaf senescence in zoysiagrass (Zoysia japonica). Plant Biotechnology Reports, 2009, 3(4): 285-292. doi: 10.1007/s11816-009-0104-9
[53] ZHANG J, XING J, LU Q, YU G, XU B, HUANG B. Transcriptional regulation of chlorophyll-catabolic genes associated with exogenous chemical effects and genotypic variations in heat-induced leaf senescence for perennial ryegrass. Environmental and Experimental Botany, 2019, 167: 103858. doi: 10.1016/j.envexpbot.2019.103858
[54] ZHANG J, SHI Y, ZHANG X, DU H, XU B, HUANG B. Melatonin suppression of heat-induced leaf senescence involves changes in abscisic acid and cytokinin biosynthesis and signaling pathways in perennial ryegrass (Lolium perenne L.). Environmental and Experimental Botany, 2017, 138: 36-45. doi: 10.1016/j.envexpbot.2017.02.012
[55] MA X, JING Z, BURGESS P, ROSSI S, HUANG B. Interactive effects of melatonin and cytokinin on alleviating drought-induced leaf senescence in creeping bentgrass (Agrostis stolonifera). Environmental and Experimental Botany, 2018, 145: 1-11. doi: 10.1016/j.envexpbot.2017.10.010
[56] GREEN P J. The ribonucleases of higher plants. Annual Review of Plant Physiology and Plant Molecular Biology, 1994, 45(1): 421-445. doi: 10.1146/annurev.pp.45.060194.002225
[57] GRAHAM I A, EASTMOND P J. Pathways of straight and branched chain fatty acid catabolism in higher plants. Progress in Lipid Research, 2002, 41(2): 156-181. doi: 10.1016/S0163-7827(01)00022-4
[58] LEVERENTZ M K, WAGSTAFF C, ROGERS H J, STEAD A D, CHANASUT U, SILKOWSKI H, THOMAS B, WEICHERT H, FEUSSNER I, GRIFFITHS G. Characterization of a novel lipoxygenase-independent senescence mechanism in Alstroemeria peruviana floral tissue. Plant Physiology, 2002, 130(1): 273-283. doi: 10.1104/pp.000919
[59] DOORN W G, BALK P, HOUWELINGEN A, HOEBERICHTS F, HALL R, VORST O, SCHOOT C, WORDRAGEN M. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology, 2003, 53(6): 845-863. doi: 10.1023/B:PLAN.0000023670.61059.1d
[60] 孙玉莹, 毕京翠, 赵志超, 程治军, 万建民. 作物叶片衰老研究进展. 作物杂志, 2013(4): 11-19. SUN Y Y, BI J C, ZHAO Z C, CHENG Z J, WAN J M. The advancement on leaf senescence in crops. Crop Journal, 2013(4): 11-19.
[61] MARTINELLI T, WHITTAKER A, MASCLAUX-DAUBRESSE C, FARRANT J M, BRILLI F, LORETO F, VAZZANA C. Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 ‘resurrection’ plant Sporobolus stapfianus during dehydration stress. Journal of Experimental Botany, 2007, 58(14): 3929-3939. doi: 10.1093/jxb/erm247
[62] HU L, ZHANG Z, XIANG Z, YANG Z. Exogenous application of citric acid ameliorates the adverse effect of heat stress in tall fescue (Lolium arundinaceum). Frontiers in Plant Science, 2016, 7: 179.
[63] KIM H, KIM H J, VU Q T, JUNG S, MCCLUNG C T, HONG S, NAM H G. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(33): 8448-8453. doi: 10.1073/pnas.1722407115
[64] WANG G, LI K, ZHOU C. Identification and functional analysis of senescence-associated genes in wheat//GUO Y. (eds). Plant Senescence. Methods in Molecular Biology. New York: Humana Press, 2018, 1744: 237-246.
[65] AY N, CLAUß K, BARTH O, HUMBECK K. Identification and characterization of novel senescence-associated genes from barley (Hordeum vulgare) primary leaves. Plant Biology, 2008, 10(s1): 121-135.
[66] SARWAT M. Leaf senescence in plants: Nutrient remobilization and gene regulation. // SARWAT M, AHMAD A, ABDIN M, IBRAHIM M. (eds). Stress Signaling in Plants: Genomics and Proteomics Perspective, Volume 2. Cham: Springer International Publishing, 2017: 301-316.
[67] UAUY C, DISTELFELD A, FAHIMA T, BLECHL A, DUBCOVSKY J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science, 2006, 314: 1298-1301. doi: 10.1126/science.1133649
[68] MA X, ZHANG Y, TUREČKOV V, XUE G P, FERNIE A R, MUELLER-ROEBER B, BALAZADEH S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiology, 2018, 177(3): 1286-1302. doi: 10.1104/pp.18.00292
[69] GU L, WEI H, WANG H, SU J, YU S. Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genetics, 2018, 19(1): 48. doi: 10.1186/s12863-018-0653-4
[70] KIM T, KANG K, KIM S H, AN G, PAEK N C. OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. International Journal of Molecular Sciences, 2019, 20(18): 4437. doi: 10.3390/ijms20184437
[71] WEILAN P, SUK-HWAN K, BYOUNG-DOO L, GYNHEUNG A, YASUHITO S, NAM-CHON P. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. Journal of Experimental Botany, 2019, 70(10): 2699-2715. doi: 10.1093/jxb/erz095
[72] PIAO W, SAKURABA Y, PAEK N C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Reports, 2019, 52(11): 653-658. doi: 10.5483/BMBRep.2019.52.11.071
[73] LI P, YANG H, LIU G, MA W, LI C, HUO H, HE J, LIU L. PpSARK regulates moss senescence and salt tolerance through ABA related pathway. International Journal of Molecular Sciences, 2018, 19(9): 2609. doi: 10.3390/ijms19092609
[74] JEHANZEB M, ZHENG X, MIAO Y. The role of the S40 gene family in leaf senescence. International Journal of Molecular Sciences, 2017, 18(10): 2152. doi: 10.3390/ijms18102152
[75] ZHENG X, JEHANZEB M, HABIBA, ZHANG Y, LI L, MIAO Y. Characterization of S40-like proteins and their roles in response to environmental cues and leaf senescence in rice. BMC Plant Biology, 2019, 19(1): 174. doi: 10.1186/s12870-019-1767-1
[76] YU T, LU X, BAI Y, MEI X, GUO Z, LIU C, CAI Y. Overexpression of the maize transcription factor ZmVQ52 accelerates leaf senescence in Arabidopsis. PLoS One, 2019, 14(8): e0221949. doi: 10.1371/journal.pone.0221949
[77] DING X, ZENG J, HUANG L, LI X, SONG S, PEI Y. Senescence-induced expression of ZmSUT1 in cotton delays leaf senescence while the seed coat-specific expression increases yield. Plant Cell Reports, 2019, 38(8): 991-1000. doi: 10.1007/s00299-019-02421-1
[78] SEKHON R S, SASKI C, KUMAR R, FLINN B S, LUO F, BEISSINGER T M, ACKERMAN A J, BREITZMAN M W, BRIDGES W C, LEON N D, KAEPPLER S M. Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. The Plant Cell, 2019, 31(9): 1968-1989. doi: 10.1105/tpc.18.00930
[79] YANG J, UDVARDI M. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. Journal of Experimental Botany, 2017, 69(4): 855-865.
[80] XU B, YU G, LI H, XIE Z, WEN W, ZHANG J, HUANG B. Knockdown of STAYGREEN in perennial ryegrass (Lolium perenne L.) leads to transcriptomic alterations related to suppressed leaf senescence and improved forage quality. Plant and Cell Physiology, 2018, 60(1): 202-212.
[81] HERATH H M S D, WEERASING A R, WIJESINGHE C R. Constructing and analyzing gene regulatory networks in leaf senescence of Arabidopsis thalina. //Seventeenth International Conference on Advances in ICT for Emerging Regions (ICTer). Colombo: ICTer, 2017: 1-8.
[82] HAQ S, KHAN A, ALI M, KHATTAK A M, GAI W X, ZHANG H X, WEI A M, GONG Z H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences, 2019, 20(21): 5321. doi: 10.3390/ijms20215321
[83] MUGA A, MORO F. Thermal adaptation of heat shock proteins. Current Protein and Peptide Science, 2008, 9(6): 552-566. doi: 10.2174/138920308786733903
[84] RAO N K S, SHIVASHANKARA K S, LAXMAN R H. Abiotic Stress Physiology of Horticultural Crops. New Delhi: Springer India, 2016.
[85] 许声涛, 孙文香, 田进平, 王崇英. 植物热激蛋白HSP100/ClpB及其在提高植物抗热性和抗寒性中的应用. 植物生理学通讯, 2008, 44(4): 197-203. XU S T, SUN W X, TIAN J P, WANG C Y. Plant heat shock protein HSP100/ClpB and its application in improving plant heat resistance and cold resistance. Plant Physiology Communications, 2008, 44(4): 197-203.
[86] WANG W, VINOCUR B, SHOSEYOV O, ALTMAN A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science, 2004, 9(5): 244-252. doi: 10.1016/j.tplants.2004.03.006
[87] GALLIE D R, GROVER A. Arabidopsis thaliana Hsp100 proteins: Kith and kin. Cell Stress and Chaperones, 2001, 6(3): 219-224. doi: 10.1379/1466-1268(2001)006<0219:ATHPKA>2.0.CO;2
[88] KEELER S J. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiology, 2000, 123(3): 1121-1132. doi: 10.1104/pp.123.3.1121
[89] GROVER A, MITTAL D, NEGI M, LAVANIA D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Science, 2013, 205/206: 38-47. doi: 10.1016/j.plantsci.2013.01.005
[90] MAKHNEVYCH T, HOURY W A. The role of Hsp90 in protein complex assembly. Biochimica et Biophysica Acta, 2012, 1823(3): 674-682. doi: 10.1016/j.bbamcr.2011.09.001
[91] JOHNSON J L, CHIOSIS G, DICKEY C A. A global view of Hsp90 functions. Nature Structural and Molecular Biology, 2013, 20(1): 1-4. doi: 10.1038/nsmb.2481
[92] KIM T S, JANG C Y, KIM H D, LEE J Y, AHN B Y, KIM J. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Molecular Biology of the Cell, 2005, 17(2): 824-833.
[93] BREIMAN A. Plant Hsp90 and its co-chaperones. Current Protein and Peptide Science, 2014, 15(3): 232-244. doi: 10.2174/1389203715666140331115603
[94] XU J, XUE C, XUE D, ZHAO J, GAI J, GUO N, XING H. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS One, 2013, 8(7): e69810. doi: 10.1371/journal.pone.0069810
[95] ZHANG M, SHEN Z, MENG G, LU Y, WANG Y. Genome-wide analysis of the Brachypodium distachyon (L.) P. Beauv. Hsp90 gene family reveals molecular evolution and expression profiling under drought and salt stresses. PLoS One, 2017, 12(12): e0189187. doi: 10.1371/journal.pone.0189187
[96] VIRDI A S, THAKUR A, DUTT S, KUMAR S, SINGH P. Asorghum 85 kDa heat stress-modulated protein shows calmodulin-binding properties and cross-reactivity to anti-Neurospora crassa Hsp 80 antibodies. FEBS Letters, 2009, 583(4): 767-770. doi: 10.1016/j.febslet.2009.01.025
[97] BREUSEGEM F, DEKEYSER R, GARCIA A B, CLAES B, GIELEN J, MONTAGU M, CAPLAN A B. Heat-inducible rice hsp82 and hsp70 are not always co-regulated. Planta, 1994, 193(1): 57-66. doi: 10.1007/BF00191607
[98] USMAN M G, RAFII M Y, MARTINI M Y, YUSUFF O A, ISMAIL M R, MIAH G. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews, 2017, 33(1): 26-39. doi: 10.1080/02648725.2017.1340546
[99] DUAN Y H, GUO J, DING K, WANG S J, ZHANG H, DAI X W, CHEN Y Y, GOVERS F, HUANG L L, KANG Z S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Molecular Biology Reports, 2011, 38(1): 301-307. doi: 10.1007/s11033-010-0108-0
[100] QI Y, WANG H, ZOU Y, LIU C, LIU Y, WANG Y, ZHANG W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. Febs Letters, 2011, 585(1): 231-239. doi: 10.1016/j.febslet.2010.11.051
[101] ONO K, HIBINO T, KOHINATA T, SUZUKI S, TANAKA Y, NAKAMURA T, TAKABE T, TAKABE T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica enhances the high-temperatue tolerance of tobacco during germination and early growth. Plant Science, 2001, 160(3): 455-461. doi: 10.1016/S0168-9452(00)00412-X
[102] SUNG D Y, GUY C L. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. evidence for pleiotropic consequences. Plant Physiology, 2003, 132(2): 979-987. doi: 10.1104/pp.102.019398
[103] ALVIM F C, CAROLINO S M, CASCARDO J C, NUNES C C, MARTINEZ C A, OTONI W C, FONTES E P. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiology, 2001, 126(3): 1042-1054. doi: 10.1104/pp.126.3.1042
[104] SUGINO M, HIBINO T, TANAKA Y, NII N, TAKABE T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica acquires resistance to salt stress in transgenic tobacco plants. Plant Science, 1999, 146(2): 81-88. doi: 10.1016/S0168-9452(99)00086-2
[105] AL-WHAIBI M H. Plant heat-shock proteins: A mini review. Journal of King Saud University Science, 2011, 23(2): 139-150. doi: 10.1016/j.jksus.2010.06.022
[106] HAQ S U, KHAN A, ALI M, GAI W X, ZHANG H X, YU Q H, YANG S B, WEI A M, GONG Z H. Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta, 2019, 250(6): 2127-2145. doi: 10.1007/s00425-019-03290-4
[107] LINDQUIST S, PARSELL D A. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annual Review of Genetics, 1993, 27(1): 437-496. doi: 10.1146/annurev.ge.27.120193.002253
[108] TOMINAGA H, COURY D A, AMANO H, MIKI W, KAKINUMA M. cDNA cloning and expression analysis of two heat shock protein genes, Hsp90 and Hsp60, from a sterile Ulva pertusa (Ulvales, Chlorophyta). Fisheries Science, 2012, 78(2): 415-429. doi: 10.1007/s12562-011-0451-7
[109] WATERS E R. The evolution, function, structure, and expression of the plant sHSPs. Journal of Experimental Botany, 2013, 642(2): 391-403.
[110] HASLBECK M, STRAUCH A. The function of small heat-shock proteins and their implication in proteostasis. Essays in Biochemistry, 2016, 60(2): 163-172. doi: 10.1042/EBC20160010
[111] DAFNY-YELIN M, TZFIRA T, VAINSTEIN A, ADAM Z. Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Molecular Biology, 2008, 67(4): 363-373. doi: 10.1007/s11103-008-9326-4
[112] MAIMBO M, OHNISHI K, HIKICHI Y, YOSHIOKA H, KIBA A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiology, 2007, 145(4): 1588-1599. doi: 10.1104/pp.107.105353
[113] 郭虹霞, 王创云, 赵丽, 王陆军, 王晋, 侯雅静, 郭晶心. 小分子热激蛋白的研究进展. 山西农业科学, 2013, 41(12): 1421-1423. GUO H X, WANG C Y, ZHAO L, WANG L J, WANG J, HOU Y J, GUO J X. Research progress of small molecule heat shock proteins. Journal of Shanxi Agricultural Sciences, 2013, 41(12): 1421-1423.
[114] SATO Y, YOKOYA S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Reports, 2008, 27(2): 329-334. doi: 10.1007/s00299-007-0470-0
[115] CUI Y, WANG M, YIN X, XU G, SONG S, LI M, LIU K, XIA X. OsMSR3, a small heat shock protein, confers enhanced tolerance to copper stress in Arabidopsis thaliana. International Journal of Molecular Sciences, 2019, 20(23): 6096. doi: 10.3390/ijms20236096
[116] REDDY P S, SHARMA K K, VADEZ V, REDDY M K. Molecular cloning and differential expression of cytosolic class I small hsp gene family in Pennisetum glaucum (L.). Applied Biochemistry and Biotechnology, 2015, 176(2): 598-612. doi: 10.1007/s12010-015-1598-y
[117] SUN X, SUN C, LI Z, HU Q, HAN L, LUO H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant, Cell and Environment, 2016, 39(6): 1320-1337. doi: 10.1111/pce.12683
[118] ZHANG Y X, ZHANG Y H, GAO H, CHAI T Y. Gene expression analysis of ubiquitin from bean under biotic and abiotic stress. Acta Botanica Boreali-occidentalia Sinica, 2002, 22(3): 505-510.
[119] BELKNAP W R, GARBARINO J E. The role of ubiquitin in plant senescence and stress responses. Trends in Plant Science, 1996, 1(10): 331-335. doi: 10.1016/S1360-1385(96)82593-0
[120] IRENE S, LAURA C, SUSANA R. Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Frontiers in Plant Science, 2018, 9: 139. doi: 10.3389/fpls.2018.00139
[121] LEE J H, KIM W T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Molecules and Cells, 2011, 31(3): 201-208. doi: 10.1007/s10059-011-0031-9
[122] YOSHID T, OHAMA N, NAKAJIMA J, KIDOKORO S, MIZOI J, NAKASHIMA K, MARUYAMA K, KIM J, SEKI M, TODAKA D, OSAKABE Y, SAKUMA Y, SCHÖFFL F, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Molecular Genetics and Genomics, 2011, 286(5/6): 321-332. doi: 10.1007/s00438-011-0647-7
[123] JACOB P, HERIBERT H, BENDAHMANE A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnology Journal, 2016, 15(4): 405-414.
[124] GUO M, LIU J H, MA X, LUO D X, GONG Z H, LU M H. The plant heat stress transcription factors (HSFs): Structure, regulation and function in response to abiotic stresses. Frontiers in Plant Science, 2016, 7: 114.
[125] SHIM D, HWANG J, LEE J, LEE S, CHOI Y, AN G, MARTIONIA E, LEE Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. The Plant Cell, 2009, 21(12): 4031-4043. doi: 10.1105/tpc.109.066902
[126] FUJIMOTO M, NAKAI A. The heat shock factor family and adaptation to proteotoxic stress. The FEBS Journal, 2010, 277(20): 4112-4125. doi: 10.1111/j.1742-4658.2010.07827.x
[127] KOSKULL-DÖRING P, SCHARF K D, NOVER L. The diversity of plant heat stress transcription factors. Trends in Plant Science, 2007, 12(10): 452-457. doi: 10.1016/j.tplants.2007.08.014
[128] 黄祥富, 黄上志, 傅家瑞. 植物热激蛋白的功能及其基因表达的调控. 植物学报, 1999, 16(5): 530. doi: 10.3969/j.issn.1674-3466.1999.05.008 HUANG X F, HUANG S Z, FU J R. Function of plant heat shock proteins and regulation of gene expression. Botany Gazette, 1999, 16(5): 530. doi: 10.3969/j.issn.1674-3466.1999.05.008
[129] NOLLEN E A A, MORIMOTO R I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing 'heat shock' proteins. Journal of Cell Science, 2002, 115(14): 2809-2816.
[130] ARRIGO A P. Small stress proteins: Chaperones that act as regulators of intracellular redox state and programmed cell death. Biological Chemistry, 1998, 379(1): 19-26.
[131] DIAMANT S, ELIAHU N, ROSENTHAL D, GOLUBINOFF P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. Journal of Biological Chemistry, 2001, 276(43): 39586. doi: 10.1074/jbc.M103081200
[132] VINER R I, CLEGG V J S. Influence of trehalose on the molecular chaperone activity of p26, a small heat shock/α-crystallin protein. Cell Stress and Chaperones, 2001, 6(2): 126-135.
[133] WANG Y, DAI Y, TAO X, WANG J Z, CHENG H Y, YANG H, MA X R. Heat shock factor genes of tall fescue and perennial ryegrass in response to temperature stress by RNA-Seq analysis. Frontiers in Plant Science, 2016, 6: 1226.
[134] RODZIEWICZ P, SWARCEWICZ B, CHMIELEWSKA K, WOJAKOWSKA A, STOBIECKI M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum, 2014, 36(1): 1-19. doi: 10.1007/s11738-013-1402-y
[135] SONG G, YUAN S, WEN X, XIE Z, LOU L, HU B, CAI Q, XU B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Reports, 2018, 37(11): 1485-1497. doi: 10.1007/s00299-018-2318-1
[136] WANG K, ZHANG X, GOATLEY M, ERVIN E. Heat shock proteins in relation to heat stress tolerance of creeping bentgrass at different N levels. PLoS One, 2014, 9(7): e102914. doi: 10.1371/journal.pone.0102914
-
表 1 热激蛋白分类及功能
Table 1 Classification and functions of heat shock proteins
热激蛋白分类
Classification of heat shock protein主要功能
Main functionHSP100 溶解变性蛋白
Dissolved denatured proteinHSP90 (83~90 kD) 促进蛋白质成熟、折叠和组装
Promotes ripening, folding assemblyHSP70 (66~78 kD) 防止聚集,协助重新折叠,蛋白质导入和易位,信号转导和转录激活
Prevents aggregation, assists in refolding, protein introduction and
translocation, signal transduction, and transcriptional activationHSP60 (60 kD) 蛋白质折叠与辅助折叠
Folding and auxiliary foldingsHSPs (15~42 kD) 防止聚合和稳定异常蛋白
Prevents polymerization and stabilizes abnormal proteins泛素 Ubiquitin (8.5 kD) 标记需要分解的蛋白,参与蛋白质降解
To label proteins that need to be broken down and participates in protein degradation -
[1] ZHU J K. Abiotic stress signaling and responses in plants. Cell, 2016, 167(2): 313-324. doi: 10.1016/j.cell.2016.08.029
[2] WANG W, VINOCUR B, ALTMAN A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta, 2003, 218(1): 1-14. doi: 10.1007/s00425-003-1105-5
[3] KHAN S A, LI M Z, WANG S M, YIN H J. Revisiting the role of plant transcription factors in the battle against abiotic stress. International Journal of Molecular Sciences, 2018, 19(6): 1634. doi: 10.3390/ijms19061634
[4] SMART C M. Gene expression during leaf senescence. New Phytologist, 2006, 126(3): 419-448.
[5] GAN S, AMASINO R M, AMASINO R M. Making sense of senescence: Molecular genetic regulation and manipulation of leaf senescence. Plant Physiology, 1997, 113(2): 313-319. doi: 10.1104/pp.113.2.313
[6] GAN S. Mitotic and postmitotic senescence in plants. Science of Aging Knowledge Environment, 2003, 2003(38): 7.
[7] NOODÉN L D, GUIAMÉT J J, JOHN I. Senescence mechanisms. Physiologia Plantarum, 2010, 101(4): 746-753.
[8] SCHARF K D, BERBERICH T, EBERSBERGER I, NOVER L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochimica Biophysica Acta, 2018, 1819(2): 104-119.
[9] REDDY P S, MALLIKARJUNA G, KAUL T, CHAKRADHAR T, MISHRE R N, SOPORY S K, REDDY M K. Molecular cloning and characterization of gene encoding for cytoplasmic Hsc70 from Pennisetum glaucum may play a protective role against abiotic stresses. Molecular Genetics and Genomics, 2010, 283(3): 243-254. doi: 10.1007/s00438-010-0518-7
[10] BOSTON R S, VIITANEN P V, VIERLING E. Molecular chaperones and protein folding in plants. Plant Molecular Biology, 1996, 32(1/2): 191-222. doi: 10.1007/BF00039383
[11] FU X, ZOU Z. Abiotic regulation: A common way for proteins to modulate their functions. Current Protein and Peptide Science, 2015, 16(3): 188-195. doi: 10.2174/1389203716666150224124429
[12] WATERS E R, LEE G J, VIERLING E. Evolution, structure and function of the small heat shock proteins in plants. Journal of Experimental Botany, 1996, 47(3): 325-338. doi: 10.1093/jxb/47.3.325
[13] KOOYERS, NICHOLAS J. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Science, 2015, 234: 155-162. doi: 10.1016/j.plantsci.2015.02.012
[14] MUNNÉ-BOSCH S, ALEGRE L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Functional Plant Biology, 2004, 31(3): 8808-8818.
[15] KRUPINSKA K. Fate and activities of plastids during leaf senescence. The Structure and Function of Plastids, 2007, 23: 433-449.
[16] GRAHAM I A, LEAVER C J, SMITH S M. Induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell, 1992, 4(3): 349-357. doi: 10.2307/3869545
[17] GILES K L, COHEN D, BEARDSELL M F. Effects of water stress on the ultrastructure of leaf cells of sorghum bicolor. Plant Physiology, 1976, 57(1): 11-14. doi: 10.1104/pp.57.1.11
[18] 杨树勋. 植物叶片衰老机理及在烟叶生产上的应用. 作物研究, 2018, 32(1): 90-96. YANG S X. Mechanisms of plant senescence and their application in tobacco leaf production. Crop Research, 2018, 32(1): 90-96.
[19] JANEČKOVÁ H, HUSIČKOVÁ A, FERRETTI U, PRČINA M, PILAROVÁ E, PLAČKOVÁ L, POSPÍŠIL P, DOLEŽAL K, ŠPUNDOVÁ M. The interplay between cytokinins and light during senescence in detached Arabidopsis leaves. Plant, Cell and Environment, 2018, 41(8): 1870-1885. doi: 10.1111/pce.13329
[20] FELLER U, KEIST M. Senescence and Nitrogen Metabolism in Annual Plants. Berlin: Springer Netherlands. 1986: 219-234.
[21] RAY S, CHOUDHURI M A. Flag leaf senescence in intact rice plant: Effects of hormones on the activities of “senescence-enzymes” during leaf age at the reproductive development. Biochemie Und Physiologie Der Pflanzen, 1980, 175(4): 346-353. doi: 10.1016/S0015-3796(80)80075-8
[22] WILSON K A, MCMANUS M T, GORDON M E, JORDAN T W. The proteomics of senescence in leaves of white clover, Trifolium repens (L.). Proteomics, 2002, 2(9): 1114-1122. doi: 10.1002/1615-9861(200209)2:9<1114::AID-PROT1114>3.0.CO;2-O
[23] MATILE P, WINKENBACH F. Function of lysosomes and lysosomal enzymes in the senescing corolla of the morning glory (Ipomoea purpurea). Journal of Experimental Botany, 1971, 22(4): 759-771. doi: 10.1093/jxb/22.4.759
[24] MARTIN C, THIMANN K V. Role of protein synthesis in the senescence of leaves: II. The influence of amino acids on senescence. Plant Physiology, 1972, 50(4): 432-437. doi: 10.1104/pp.50.4.432
[25] 宋松泉, 苏卫珍, 彭晓南. 杂交水稻离体叶片衰老与蛋白质代谢的关系. 逻辑学研究, 1995(1): 20-23. SONG S Q, SU W Z, PENG X N. Relationship between leaf senescence and protein metabolism in vitro of hybrid rice. Studies in Logic, 1995(1): 20-23.
[26] GLICKMAN M H, CIECHANOVER A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews, 2002, 82(2): 373-428. doi: 10.1152/physrev.00027.2001
[27] JESPERSEN D, ZHANG J, HUANG B. Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Science, 2016, 249: 1-12. doi: 10.1016/j.plantsci.2016.04.016
[28] JESPERSEN D, XU C, HUANG B C. Membrane proteins associated with heat‐induced leaf senescence in a cool‐season grass species. Crop Science, 2015, 55(2): 837-850. doi: 10.2135/cropsci2014.04.0335
[29] YAN C S, LI D Q, ZHANG J H. Plant leaf senescence and oxidative stress. Chinese Bulletin of Botany, 2005, 10(3): 515-534.
[30] HURA T, HURA K, OSTROWSKA A, GADZINOWSKA J, FIUST A. Water stress-induced flag leaf senescence may be accelerated by rehydration. Journal of Plant Physiology, 2019, 236: 109-116. doi: 10.1016/j.jplph.2019.01.013
[31] MITTLER G, SUZUKI N, CIFTCI-YILMAZ S, MITTLER R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell and Environment, 2010, 33(4): 453-467. doi: 10.1111/j.1365-3040.2009.02041.x
[32] TSENG M J, LIU C W, YIU J C. Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiology and Biochemistry, 2007, 45(10/11): 822-833. doi: 10.1016/j.plaphy.2007.07.011
[33] LEE S, SEO P J, LEE H J, PARK C M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant Journal, 2012, 70(5): 831-844. doi: 10.1111/j.1365-313X.2012.04932.x
[34] ZHANG J, LI H, XU B, LI J, HUANG B. Exogenous melatonin suppresses dark-induced leaf senescence by activating the superoxide dismutase-catalase antioxidant pathway and down-regulating chlorophyll degradation in excised leaves of perennial ryegrass (Lolium perenne L.). Frontiers in Plant Science, 2016, 7: 1500.
[35] JIBRAN R, HUNTER D, DIJKWEL P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Molecular Biology, 2013, 82(6): 547-561. doi: 10.1007/s11103-013-0043-2
[36] ZHAO Y, CHAN Z, GAO J, XING L, CAO M, YU C, HU Y, YOU J, SHI H, ZHU Y, GONG Y, MU Z, WANG H, DENG X, WANG P, BRESSAN R A, ZHU J K. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(7): 1949-1954. doi: 10.1073/pnas.1522840113
[37] MANOHARAN K, PRASAD R, GUHA-MUKHERJEE S. Senescence-related lipid changes in callus cultures of Datura innoxia. Phytochemistry, 1990, 29(8): 2529-2531. doi: 10.1016/0031-9422(90)85181-E
[38] DONG H, NIU Y, LI W, ZHANG D. Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. Journal of Experimental Botany, 2008, 59(6): 1295-1304. doi: 10.1093/jxb/ern035
[39] HE Y, ZHANG Z, LI L, TANG S, WU J L. Genetic and physio-biochemical characterization of a novel premature senescence leaf mutant in rice (Oryza sativa L.). International Journal of Molecular Sciences, 2018, 19(8): 2339. doi: 10.3390/ijms19082339
[40] XU B, LI H, YU G, ZHANG J, HUANG B. Characterization and transcriptional regulation of chlorophyll b reductase gene NON-YELLOW COLORING 1 associated with leaf senescence in perennial ryegrass (Lolium perenne L.). Environmental and Experimental Botany, 2018, 149(1): 43-50.
[41] JING Z, YU G, WEN W, MA X, HUANG B. Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. Journal of Experimental Botany, 2015, 67(3): 935-945.
[42] OH S A, PARK J H, LEE G I, PAEK K H, PARK S K, NAM H G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. The Plant Journal, 2003, 12(3): 527-535.
[43] JOHN I, DRAKE R, FARRELL A, COOPER W, LEE P, HORTON P, GRIERSON D. Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: Molecular and physiological analysis. The Plant Journal, 1995, 7: 483-490. doi: 10.1046/j.1365-313X.1995.7030483.x
[44] IRIGOYEN J J, EMERICH D W, SÁNCHEZ-DÍAZ M. Alfalfa leaf senescence induced by drought stress: Photosynthesis, hydrogen peroxide metabolism, lipid peroxidation and ethylene evolution. Physiologia Plantarum, 2006, 84(1): 67-72.
[45] SCHNEIDER H M, TOBIAS W, POSTMA J A, BROWN K M, LYNCH J P. Ethylene modulates root cortical senescence in barley. Annals of Botany, 2018, 122(1): 95-105. doi: 10.1093/aob/mcy059
[46] GAN S, AMASINO R M. Cytokinins in plant senescence: From spray and pray to clone and play. Bioessays, 1996, 18(7): 557-565. doi: 10.1002/bies.950180707
[47] TALLA S K, PANIGRAPHY M, KAPPARA S, NIROSHA P, NEELAMRAJU S, RAMANAN R. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. Journal of Experimental Botany, 2016, 67(6): 1839-1851. doi: 10.1093/jxb/erv575
[48] MA X, ZHANG J, HUANG B. Cytokinin-mitigation of salt-induced leaf senescence in perennial ryegrass involving the activation of antioxidant systems and ionic balance. Environmental and Experimental Botany, 2016, 125: 1-11. doi: 10.1016/j.envexpbot.2016.01.002
[49] MARKOVICH O, STEINER E, KOURIL Š, TARKOWSKI P, AHARONI A, ELBAUM R. Silicon promotes cytokinin biosynthesis and delays senescence in Arabidopsis and Sorghum. Plant, Cell and Environment, 2017, 40(7): 1189-1196. doi: 10.1111/pce.12913
[50] JESPERSEN D, YU J, HUANG B. Metabolite responses to exogenous application of nitrogen, cytokinin, and ethylene inhibitors in relation to heat-induced senescence in creeping bentgrass. PLoS One, 2015, 10(3): e0123744. doi: 10.1371/journal.pone.0123744
[51] GUO P, LI Z, HUANG P, LI B, FANG S, CHU J, GUO H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. The Plant Cell, 2017, 29(11): 2854-2870. doi: 10.1105/tpc.17.00438
[52] CHENG X, DAI X, ZENG H, LI Y, WEI T, HAN L. Gene expression involved in dark-induced leaf senescence in zoysiagrass (Zoysia japonica). Plant Biotechnology Reports, 2009, 3(4): 285-292. doi: 10.1007/s11816-009-0104-9
[53] ZHANG J, XING J, LU Q, YU G, XU B, HUANG B. Transcriptional regulation of chlorophyll-catabolic genes associated with exogenous chemical effects and genotypic variations in heat-induced leaf senescence for perennial ryegrass. Environmental and Experimental Botany, 2019, 167: 103858. doi: 10.1016/j.envexpbot.2019.103858
[54] ZHANG J, SHI Y, ZHANG X, DU H, XU B, HUANG B. Melatonin suppression of heat-induced leaf senescence involves changes in abscisic acid and cytokinin biosynthesis and signaling pathways in perennial ryegrass (Lolium perenne L.). Environmental and Experimental Botany, 2017, 138: 36-45. doi: 10.1016/j.envexpbot.2017.02.012
[55] MA X, JING Z, BURGESS P, ROSSI S, HUANG B. Interactive effects of melatonin and cytokinin on alleviating drought-induced leaf senescence in creeping bentgrass (Agrostis stolonifera). Environmental and Experimental Botany, 2018, 145: 1-11. doi: 10.1016/j.envexpbot.2017.10.010
[56] GREEN P J. The ribonucleases of higher plants. Annual Review of Plant Physiology and Plant Molecular Biology, 1994, 45(1): 421-445. doi: 10.1146/annurev.pp.45.060194.002225
[57] GRAHAM I A, EASTMOND P J. Pathways of straight and branched chain fatty acid catabolism in higher plants. Progress in Lipid Research, 2002, 41(2): 156-181. doi: 10.1016/S0163-7827(01)00022-4
[58] LEVERENTZ M K, WAGSTAFF C, ROGERS H J, STEAD A D, CHANASUT U, SILKOWSKI H, THOMAS B, WEICHERT H, FEUSSNER I, GRIFFITHS G. Characterization of a novel lipoxygenase-independent senescence mechanism in Alstroemeria peruviana floral tissue. Plant Physiology, 2002, 130(1): 273-283. doi: 10.1104/pp.000919
[59] DOORN W G, BALK P, HOUWELINGEN A, HOEBERICHTS F, HALL R, VORST O, SCHOOT C, WORDRAGEN M. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology, 2003, 53(6): 845-863. doi: 10.1023/B:PLAN.0000023670.61059.1d
[60] 孙玉莹, 毕京翠, 赵志超, 程治军, 万建民. 作物叶片衰老研究进展. 作物杂志, 2013(4): 11-19. SUN Y Y, BI J C, ZHAO Z C, CHENG Z J, WAN J M. The advancement on leaf senescence in crops. Crop Journal, 2013(4): 11-19.
[61] MARTINELLI T, WHITTAKER A, MASCLAUX-DAUBRESSE C, FARRANT J M, BRILLI F, LORETO F, VAZZANA C. Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 ‘resurrection’ plant Sporobolus stapfianus during dehydration stress. Journal of Experimental Botany, 2007, 58(14): 3929-3939. doi: 10.1093/jxb/erm247
[62] HU L, ZHANG Z, XIANG Z, YANG Z. Exogenous application of citric acid ameliorates the adverse effect of heat stress in tall fescue (Lolium arundinaceum). Frontiers in Plant Science, 2016, 7: 179.
[63] KIM H, KIM H J, VU Q T, JUNG S, MCCLUNG C T, HONG S, NAM H G. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(33): 8448-8453. doi: 10.1073/pnas.1722407115
[64] WANG G, LI K, ZHOU C. Identification and functional analysis of senescence-associated genes in wheat//GUO Y. (eds). Plant Senescence. Methods in Molecular Biology. New York: Humana Press, 2018, 1744: 237-246.
[65] AY N, CLAUß K, BARTH O, HUMBECK K. Identification and characterization of novel senescence-associated genes from barley (Hordeum vulgare) primary leaves. Plant Biology, 2008, 10(s1): 121-135.
[66] SARWAT M. Leaf senescence in plants: Nutrient remobilization and gene regulation. // SARWAT M, AHMAD A, ABDIN M, IBRAHIM M. (eds). Stress Signaling in Plants: Genomics and Proteomics Perspective, Volume 2. Cham: Springer International Publishing, 2017: 301-316.
[67] UAUY C, DISTELFELD A, FAHIMA T, BLECHL A, DUBCOVSKY J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science, 2006, 314: 1298-1301. doi: 10.1126/science.1133649
[68] MA X, ZHANG Y, TUREČKOV V, XUE G P, FERNIE A R, MUELLER-ROEBER B, BALAZADEH S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiology, 2018, 177(3): 1286-1302. doi: 10.1104/pp.18.00292
[69] GU L, WEI H, WANG H, SU J, YU S. Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genetics, 2018, 19(1): 48. doi: 10.1186/s12863-018-0653-4
[70] KIM T, KANG K, KIM S H, AN G, PAEK N C. OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. International Journal of Molecular Sciences, 2019, 20(18): 4437. doi: 10.3390/ijms20184437
[71] WEILAN P, SUK-HWAN K, BYOUNG-DOO L, GYNHEUNG A, YASUHITO S, NAM-CHON P. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. Journal of Experimental Botany, 2019, 70(10): 2699-2715. doi: 10.1093/jxb/erz095
[72] PIAO W, SAKURABA Y, PAEK N C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Reports, 2019, 52(11): 653-658. doi: 10.5483/BMBRep.2019.52.11.071
[73] LI P, YANG H, LIU G, MA W, LI C, HUO H, HE J, LIU L. PpSARK regulates moss senescence and salt tolerance through ABA related pathway. International Journal of Molecular Sciences, 2018, 19(9): 2609. doi: 10.3390/ijms19092609
[74] JEHANZEB M, ZHENG X, MIAO Y. The role of the S40 gene family in leaf senescence. International Journal of Molecular Sciences, 2017, 18(10): 2152. doi: 10.3390/ijms18102152
[75] ZHENG X, JEHANZEB M, HABIBA, ZHANG Y, LI L, MIAO Y. Characterization of S40-like proteins and their roles in response to environmental cues and leaf senescence in rice. BMC Plant Biology, 2019, 19(1): 174. doi: 10.1186/s12870-019-1767-1
[76] YU T, LU X, BAI Y, MEI X, GUO Z, LIU C, CAI Y. Overexpression of the maize transcription factor ZmVQ52 accelerates leaf senescence in Arabidopsis. PLoS One, 2019, 14(8): e0221949. doi: 10.1371/journal.pone.0221949
[77] DING X, ZENG J, HUANG L, LI X, SONG S, PEI Y. Senescence-induced expression of ZmSUT1 in cotton delays leaf senescence while the seed coat-specific expression increases yield. Plant Cell Reports, 2019, 38(8): 991-1000. doi: 10.1007/s00299-019-02421-1
[78] SEKHON R S, SASKI C, KUMAR R, FLINN B S, LUO F, BEISSINGER T M, ACKERMAN A J, BREITZMAN M W, BRIDGES W C, LEON N D, KAEPPLER S M. Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. The Plant Cell, 2019, 31(9): 1968-1989. doi: 10.1105/tpc.18.00930
[79] YANG J, UDVARDI M. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. Journal of Experimental Botany, 2017, 69(4): 855-865.
[80] XU B, YU G, LI H, XIE Z, WEN W, ZHANG J, HUANG B. Knockdown of STAYGREEN in perennial ryegrass (Lolium perenne L.) leads to transcriptomic alterations related to suppressed leaf senescence and improved forage quality. Plant and Cell Physiology, 2018, 60(1): 202-212.
[81] HERATH H M S D, WEERASING A R, WIJESINGHE C R. Constructing and analyzing gene regulatory networks in leaf senescence of Arabidopsis thalina. //Seventeenth International Conference on Advances in ICT for Emerging Regions (ICTer). Colombo: ICTer, 2017: 1-8.
[82] HAQ S, KHAN A, ALI M, KHATTAK A M, GAI W X, ZHANG H X, WEI A M, GONG Z H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences, 2019, 20(21): 5321. doi: 10.3390/ijms20215321
[83] MUGA A, MORO F. Thermal adaptation of heat shock proteins. Current Protein and Peptide Science, 2008, 9(6): 552-566. doi: 10.2174/138920308786733903
[84] RAO N K S, SHIVASHANKARA K S, LAXMAN R H. Abiotic Stress Physiology of Horticultural Crops. New Delhi: Springer India, 2016.
[85] 许声涛, 孙文香, 田进平, 王崇英. 植物热激蛋白HSP100/ClpB及其在提高植物抗热性和抗寒性中的应用. 植物生理学通讯, 2008, 44(4): 197-203. XU S T, SUN W X, TIAN J P, WANG C Y. Plant heat shock protein HSP100/ClpB and its application in improving plant heat resistance and cold resistance. Plant Physiology Communications, 2008, 44(4): 197-203.
[86] WANG W, VINOCUR B, SHOSEYOV O, ALTMAN A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science, 2004, 9(5): 244-252. doi: 10.1016/j.tplants.2004.03.006
[87] GALLIE D R, GROVER A. Arabidopsis thaliana Hsp100 proteins: Kith and kin. Cell Stress and Chaperones, 2001, 6(3): 219-224. doi: 10.1379/1466-1268(2001)006<0219:ATHPKA>2.0.CO;2
[88] KEELER S J. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiology, 2000, 123(3): 1121-1132. doi: 10.1104/pp.123.3.1121
[89] GROVER A, MITTAL D, NEGI M, LAVANIA D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Science, 2013, 205/206: 38-47. doi: 10.1016/j.plantsci.2013.01.005
[90] MAKHNEVYCH T, HOURY W A. The role of Hsp90 in protein complex assembly. Biochimica et Biophysica Acta, 2012, 1823(3): 674-682. doi: 10.1016/j.bbamcr.2011.09.001
[91] JOHNSON J L, CHIOSIS G, DICKEY C A. A global view of Hsp90 functions. Nature Structural and Molecular Biology, 2013, 20(1): 1-4. doi: 10.1038/nsmb.2481
[92] KIM T S, JANG C Y, KIM H D, LEE J Y, AHN B Y, KIM J. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Molecular Biology of the Cell, 2005, 17(2): 824-833.
[93] BREIMAN A. Plant Hsp90 and its co-chaperones. Current Protein and Peptide Science, 2014, 15(3): 232-244. doi: 10.2174/1389203715666140331115603
[94] XU J, XUE C, XUE D, ZHAO J, GAI J, GUO N, XING H. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS One, 2013, 8(7): e69810. doi: 10.1371/journal.pone.0069810
[95] ZHANG M, SHEN Z, MENG G, LU Y, WANG Y. Genome-wide analysis of the Brachypodium distachyon (L.) P. Beauv. Hsp90 gene family reveals molecular evolution and expression profiling under drought and salt stresses. PLoS One, 2017, 12(12): e0189187. doi: 10.1371/journal.pone.0189187
[96] VIRDI A S, THAKUR A, DUTT S, KUMAR S, SINGH P. Asorghum 85 kDa heat stress-modulated protein shows calmodulin-binding properties and cross-reactivity to anti-Neurospora crassa Hsp 80 antibodies. FEBS Letters, 2009, 583(4): 767-770. doi: 10.1016/j.febslet.2009.01.025
[97] BREUSEGEM F, DEKEYSER R, GARCIA A B, CLAES B, GIELEN J, MONTAGU M, CAPLAN A B. Heat-inducible rice hsp82 and hsp70 are not always co-regulated. Planta, 1994, 193(1): 57-66. doi: 10.1007/BF00191607
[98] USMAN M G, RAFII M Y, MARTINI M Y, YUSUFF O A, ISMAIL M R, MIAH G. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews, 2017, 33(1): 26-39. doi: 10.1080/02648725.2017.1340546
[99] DUAN Y H, GUO J, DING K, WANG S J, ZHANG H, DAI X W, CHEN Y Y, GOVERS F, HUANG L L, KANG Z S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Molecular Biology Reports, 2011, 38(1): 301-307. doi: 10.1007/s11033-010-0108-0
[100] QI Y, WANG H, ZOU Y, LIU C, LIU Y, WANG Y, ZHANG W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. Febs Letters, 2011, 585(1): 231-239. doi: 10.1016/j.febslet.2010.11.051
[101] ONO K, HIBINO T, KOHINATA T, SUZUKI S, TANAKA Y, NAKAMURA T, TAKABE T, TAKABE T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica enhances the high-temperatue tolerance of tobacco during germination and early growth. Plant Science, 2001, 160(3): 455-461. doi: 10.1016/S0168-9452(00)00412-X
[102] SUNG D Y, GUY C L. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. evidence for pleiotropic consequences. Plant Physiology, 2003, 132(2): 979-987. doi: 10.1104/pp.102.019398
[103] ALVIM F C, CAROLINO S M, CASCARDO J C, NUNES C C, MARTINEZ C A, OTONI W C, FONTES E P. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiology, 2001, 126(3): 1042-1054. doi: 10.1104/pp.126.3.1042
[104] SUGINO M, HIBINO T, TANAKA Y, NII N, TAKABE T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica acquires resistance to salt stress in transgenic tobacco plants. Plant Science, 1999, 146(2): 81-88. doi: 10.1016/S0168-9452(99)00086-2
[105] AL-WHAIBI M H. Plant heat-shock proteins: A mini review. Journal of King Saud University Science, 2011, 23(2): 139-150. doi: 10.1016/j.jksus.2010.06.022
[106] HAQ S U, KHAN A, ALI M, GAI W X, ZHANG H X, YU Q H, YANG S B, WEI A M, GONG Z H. Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta, 2019, 250(6): 2127-2145. doi: 10.1007/s00425-019-03290-4
[107] LINDQUIST S, PARSELL D A. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annual Review of Genetics, 1993, 27(1): 437-496. doi: 10.1146/annurev.ge.27.120193.002253
[108] TOMINAGA H, COURY D A, AMANO H, MIKI W, KAKINUMA M. cDNA cloning and expression analysis of two heat shock protein genes, Hsp90 and Hsp60, from a sterile Ulva pertusa (Ulvales, Chlorophyta). Fisheries Science, 2012, 78(2): 415-429. doi: 10.1007/s12562-011-0451-7
[109] WATERS E R. The evolution, function, structure, and expression of the plant sHSPs. Journal of Experimental Botany, 2013, 642(2): 391-403.
[110] HASLBECK M, STRAUCH A. The function of small heat-shock proteins and their implication in proteostasis. Essays in Biochemistry, 2016, 60(2): 163-172. doi: 10.1042/EBC20160010
[111] DAFNY-YELIN M, TZFIRA T, VAINSTEIN A, ADAM Z. Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Molecular Biology, 2008, 67(4): 363-373. doi: 10.1007/s11103-008-9326-4
[112] MAIMBO M, OHNISHI K, HIKICHI Y, YOSHIOKA H, KIBA A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiology, 2007, 145(4): 1588-1599. doi: 10.1104/pp.107.105353
[113] 郭虹霞, 王创云, 赵丽, 王陆军, 王晋, 侯雅静, 郭晶心. 小分子热激蛋白的研究进展. 山西农业科学, 2013, 41(12): 1421-1423. GUO H X, WANG C Y, ZHAO L, WANG L J, WANG J, HOU Y J, GUO J X. Research progress of small molecule heat shock proteins. Journal of Shanxi Agricultural Sciences, 2013, 41(12): 1421-1423.
[114] SATO Y, YOKOYA S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Reports, 2008, 27(2): 329-334. doi: 10.1007/s00299-007-0470-0
[115] CUI Y, WANG M, YIN X, XU G, SONG S, LI M, LIU K, XIA X. OsMSR3, a small heat shock protein, confers enhanced tolerance to copper stress in Arabidopsis thaliana. International Journal of Molecular Sciences, 2019, 20(23): 6096. doi: 10.3390/ijms20236096
[116] REDDY P S, SHARMA K K, VADEZ V, REDDY M K. Molecular cloning and differential expression of cytosolic class I small hsp gene family in Pennisetum glaucum (L.). Applied Biochemistry and Biotechnology, 2015, 176(2): 598-612. doi: 10.1007/s12010-015-1598-y
[117] SUN X, SUN C, LI Z, HU Q, HAN L, LUO H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant, Cell and Environment, 2016, 39(6): 1320-1337. doi: 10.1111/pce.12683
[118] ZHANG Y X, ZHANG Y H, GAO H, CHAI T Y. Gene expression analysis of ubiquitin from bean under biotic and abiotic stress. Acta Botanica Boreali-occidentalia Sinica, 2002, 22(3): 505-510.
[119] BELKNAP W R, GARBARINO J E. The role of ubiquitin in plant senescence and stress responses. Trends in Plant Science, 1996, 1(10): 331-335. doi: 10.1016/S1360-1385(96)82593-0
[120] IRENE S, LAURA C, SUSANA R. Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Frontiers in Plant Science, 2018, 9: 139. doi: 10.3389/fpls.2018.00139
[121] LEE J H, KIM W T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Molecules and Cells, 2011, 31(3): 201-208. doi: 10.1007/s10059-011-0031-9
[122] YOSHID T, OHAMA N, NAKAJIMA J, KIDOKORO S, MIZOI J, NAKASHIMA K, MARUYAMA K, KIM J, SEKI M, TODAKA D, OSAKABE Y, SAKUMA Y, SCHÖFFL F, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Molecular Genetics and Genomics, 2011, 286(5/6): 321-332. doi: 10.1007/s00438-011-0647-7
[123] JACOB P, HERIBERT H, BENDAHMANE A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnology Journal, 2016, 15(4): 405-414.
[124] GUO M, LIU J H, MA X, LUO D X, GONG Z H, LU M H. The plant heat stress transcription factors (HSFs): Structure, regulation and function in response to abiotic stresses. Frontiers in Plant Science, 2016, 7: 114.
[125] SHIM D, HWANG J, LEE J, LEE S, CHOI Y, AN G, MARTIONIA E, LEE Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. The Plant Cell, 2009, 21(12): 4031-4043. doi: 10.1105/tpc.109.066902
[126] FUJIMOTO M, NAKAI A. The heat shock factor family and adaptation to proteotoxic stress. The FEBS Journal, 2010, 277(20): 4112-4125. doi: 10.1111/j.1742-4658.2010.07827.x
[127] KOSKULL-DÖRING P, SCHARF K D, NOVER L. The diversity of plant heat stress transcription factors. Trends in Plant Science, 2007, 12(10): 452-457. doi: 10.1016/j.tplants.2007.08.014
[128] 黄祥富, 黄上志, 傅家瑞. 植物热激蛋白的功能及其基因表达的调控. 植物学报, 1999, 16(5): 530. doi: 10.3969/j.issn.1674-3466.1999.05.008 HUANG X F, HUANG S Z, FU J R. Function of plant heat shock proteins and regulation of gene expression. Botany Gazette, 1999, 16(5): 530. doi: 10.3969/j.issn.1674-3466.1999.05.008
[129] NOLLEN E A A, MORIMOTO R I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing 'heat shock' proteins. Journal of Cell Science, 2002, 115(14): 2809-2816.
[130] ARRIGO A P. Small stress proteins: Chaperones that act as regulators of intracellular redox state and programmed cell death. Biological Chemistry, 1998, 379(1): 19-26.
[131] DIAMANT S, ELIAHU N, ROSENTHAL D, GOLUBINOFF P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. Journal of Biological Chemistry, 2001, 276(43): 39586. doi: 10.1074/jbc.M103081200
[132] VINER R I, CLEGG V J S. Influence of trehalose on the molecular chaperone activity of p26, a small heat shock/α-crystallin protein. Cell Stress and Chaperones, 2001, 6(2): 126-135.
[133] WANG Y, DAI Y, TAO X, WANG J Z, CHENG H Y, YANG H, MA X R. Heat shock factor genes of tall fescue and perennial ryegrass in response to temperature stress by RNA-Seq analysis. Frontiers in Plant Science, 2016, 6: 1226.
[134] RODZIEWICZ P, SWARCEWICZ B, CHMIELEWSKA K, WOJAKOWSKA A, STOBIECKI M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum, 2014, 36(1): 1-19. doi: 10.1007/s11738-013-1402-y
[135] SONG G, YUAN S, WEN X, XIE Z, LOU L, HU B, CAI Q, XU B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Reports, 2018, 37(11): 1485-1497. doi: 10.1007/s00299-018-2318-1
[136] WANG K, ZHANG X, GOATLEY M, ERVIN E. Heat shock proteins in relation to heat stress tolerance of creeping bentgrass at different N levels. PLoS One, 2014, 9(7): e102914. doi: 10.1371/journal.pone.0102914
-
期刊类型引用(8)
1. 杨清,刘胜红,黄二宾,杜嵘宇,王芳,邓佳. 经羧甲基壳聚糖诱导的葡萄柚果实转录组WRKY基因分析及抗性相关基因挖掘. 浙江农业学报. 2023(03): 598-614 .  百度学术
百度学术
2. 李奇勋,张敏,贾淼,李玉申,王红婷. 贮前热水处理对低温贮藏西葫芦不同组织抗冷性和抗氧化系统的影响. 安徽农业大学学报. 2023(02): 235-242 .  百度学术
百度学术
3. 宋奇琦,张小秋,宋修鹏,颜梅新,王泽平,雷敬超,黄冬梅,李秋芳,梁永检. 甘蔗HSP20基因克隆、原核表达及逆境胁迫响应. 植物生理学报. 2022(02): 371-380 .  百度学术
百度学术
4. Fei TONG,Jiwei LIU,Yuanna CHENG,Ting YE,Zhaodong LIU,Ni ZHAN,Hui YE,Liquan WU,Juan LI. Expression Analysis of Heat Shock Protein 70 Gene in Rice (Oryza sativa L.). Agricultural Biotechnology. 2022(01): 1-4+21 .  必应学术
必应学术
5. 吕宇婧,吴丹丹,孔春艳,杨宇,龚明. 小桐子Hsp70基因家族和相应miRNAs的鉴定与互作分析及其在低温适应中的作用. 植物生理学报. 2022(07): 1221-1235 .  百度学术
百度学术
6. 任玉玲,吴云,赵艳,孙胜男,赵成周,李萍. 镉胁迫诱导的两种青稞叶片衰老差异的生理学比较. 分子植物育种. 2022(22): 7574-7583 .  百度学术
百度学术
7. 朱丹,秦亚东,李林华,高焕. 高温胁迫诱导薄荷差异表达基因的分析. 安徽科技学院学报. 2022(06): 54-62 .  百度学术
百度学术
8. 毕银丽,薛子可. 丛枝菌根真菌提高植物高温胁迫抗逆性及在矿区生态修复应用展望. 中国科学基金. 2021(06): 933-939 .  百度学术
百度学术
其他类型引用(16)



 下载:
下载:


