玉米B3基因家族全基因组鉴定及表达模式
B3基因家族是植物中特有的转录因子家族,在植物生长发育及胁迫响应中有重要作用。为探究B3基因家族在玉米(Zea mays)中的基本信息及生长发育和逆境胁迫中的生物学功能,本研究对玉米全基因组B3家族成员进行鉴定并分析相关信息,依据转录组数据分析B3基因家族组织特异性表达及胁迫处理下的表达响应。结果显示,玉米B3基因家族包含88个成员,分为ABI、ARF、HSI、RAV和REM 5个亚家族,均含有B3结构域,同一个亚族成员的基因结构及保守基序(数量、类型)相似。分析发现,B3成员基因的启动子区域中均富含激素及逆境响应等功能元件。组织特异性表达分析结果显示,大部分B3基因在分生组织中优势表达。对B3家族成员在冷、热、盐、紫外和干旱这5种胁迫因子的响应表达分析显示,5个亚家族成员均可不同程度参与响应冷、热、盐、紫外和干旱胁迫因子,其中HSI亚家族中的所有成员均参与了对紫外胁迫因子的响应途径。以上结果阐明了B3基因家族在玉米全基因组中的分布、结构和系统发育特征,并依据玉米转录组数据,初步推测其在生长发育和胁迫应激响应中发挥重要作用,为深入研究B3基因家族在玉米中的功能与分子机制提供了数据基础。
English
-
B3家族是一类植物特有的转录因子家族,从单细胞绿藻到真核生物中均有发现[1],广泛分布于植物基因组中,其所有成员都具有一个长度约110个氨基酸的B3结构域,该结构域具有序列特异性DNA结合活性,最初在玉米(Zea mays)中被鉴定,命名为VIVIPAROUS1 (VP1);随后在拟南芥(Arabidopsis thaliana)中也被发现,鉴定为ABSCISIC ACID INSENSITIVE 3 (ABI)[2-3]。B3家族成员的数量因植物而异,在拟南芥、水稻(Oryza sativa)、大豆(Glycine max)中分别有118、91、108个成员[4]。根据蛋白质的结构和功能,植物中的B3家族可分为5个亚家族:RAV (related to ABI3/VP1)家族、ARF (auxin response factor)家族、ABI (abscisic acid insensitive 3)家族、REM (reproductive meristem)家族和HSI (high level expression of sugar inducible)家族[5]。在拟南芥中,ABI (一个B3结构域)、HSI (一个B3结构域和zf CW结构域)、ARF (一个B3结构域、生长素响应因子结构域和AUX/IAA结构域)、RAV (一个B3结构域和AP2结构域)和REM (多个B3结构域)具有不同的典型结构域[6]。B3的每个亚群在植物生长、发育和胁迫反应中发挥着不同的作用[7-8]。例如,ARFs在花和叶的发育、维管组织分化和根的形成中具有功能[9-10]。在拟南芥中,arf1或arf2突变体表现出发育延迟,包括开花起始和莲座叶衰老[11],而双突变体arf7arf19影响根系生长和叶片扩展[12]。目前对RAV家族的研究很少,主要集中在对种子萌发、生长发育和应激反应的调控方面[13-16]。在棉花(Gossypium hirsutum)中,过表达ABI3基因可以提高植株抗旱性及光合效率[17]。LEC2诱导体细胞胚胎形成,促进种子贮藏蛋白质和油体的积累[18]。作为一种转录抑制因子,HSIs具有抑制种子成熟基因表达的功能[19]。拟南芥中HSI2通过MSI1抑制AGL15 (与种子发育有关),从而调节种子成熟[20]。大量研究表明,RAVs与胁迫反应有关,拟南芥中过表达RAV1基因降低了对盐和干旱胁迫的适应性[21]。从另一个角度来看,TEM (TEMPRANILLO)基因(RAV家族成员)负调控开花基因FT (flowering locus T)的表达,从而抑制开花[22]。AtREM34是第1个被鉴定的REM家族成员,VRN1 (VERNALIZATION1/REM5)是第1个被鉴定与春化和促进开花相关的功能因子[23]。在拟南芥中,同时沉默REM34和REM35会影响其雌配子体和雄配子体的发育,表明REMs在开花中发挥作用[24]。
1. 材料与方法
1.1 玉米B3基因家族鉴定
玉米基因序列和蛋白序列下载自玉米Maize GDB数据库(Zm-B73-REFERENCE-NAM-5.0;https://www.maizegdb.org)。从拟南芥基因组数据库(TAIR;https://www.arabidopsis.org/)下载拟南芥B3家族蛋白序列进行blastp同源比对。在Pfam数据库(http://pfam.xfam.org/)下载B3结构域(PF02362)文件,利用HMMER软件搜索,参数设置为1 e−5,候选序列合并去重。通过PFAM (http://pfam.xfam.org/)和NCBI-CDD (https://www.ncbi.nlm.nih.gov/)工具进行结构域鉴定。利用在线工具对B3基因家族成员进行理化性质(ExPASy;https://web.expasy.org/)、跨膜结构(TMHMM;http://www.cbs.dtu.dk/)、信号肽(Signa;http://www.cbs.dtu.dk/)和亚细胞定位(CELLO;http://cello.life.nctu.edu.tw/)预测分析[25]。
1.2 玉米B3基因家族种内聚类、保守基序及基因结构分析
利用MEME9 (https://meme-suite.org/)工具对玉米B3蛋白序列进行保守基序的检索。种内聚类采用MEGA10中muscle算法进行比对,邻接法(neighbor-joining, NJ)建树。从基因结构注释文件中提取玉米B3基因的基因结构信息,并基于PFAM和NCBI-CDD数据库序列结构域推断结果,利用TBtools绘制基因结构图。
1.3 玉米及拟南芥、水稻B3基因家族系统发育树的构建
TAIR下载拟南芥蛋白序列,NCBI下载水稻蛋白序列。通过MEGA 7.0软件,对拟南芥B3基因家族、水稻B3基因家族和玉米B3基因家族蛋白序列进行多序列比对,以NJ法构建同源蛋白无根进化树,利用Figtree进行绘制。
1.4 启动子顺式作用元件分析
利用TBtools软件截获玉米B3基因上游2 000 bp的启动子序列,采用PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)分析顺式调控元件。
1.5 玉米B3基因家族基因的表达模式分析
在NCBI网站GEO公共数据库(https://www.ncbi.nlm.nih.gov/sra)中下载玉米基因芯片[26-27],利用GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/)程序进行探针匹配,提取玉米B3基因家族的组织表达数据。在SRA公共数据库(https://www.ncbi.nlm.nih.gov/geo/)下载玉米冷胁迫、热胁迫、盐胁迫、紫外胁迫和干旱胁迫的转录组数据[28-29],利用Sratools软件将转录组数据转换为fastq格式,利用fastqc进行质量控制,fastp进行数据过滤,计算每个基因的TPM值,以TPM > 1作为筛选表达的筛选标准,对转录组数据进行log2 (1 + TPM)均一化处理,并进行可视化。
2. 结果
2.1 玉米B3家族成员鉴定
利用HMMER和Blastp在玉米基因组中进行搜索得到候选B3家族成员,去除不含B3保守结构域的基因,最终获得88个ZmB3家族成员,包括4个RAV成员、40个ARF成员、37个REM成员、4个HSI3成员和3个ABI成员(表1)。并依据染色体位置和保守结构域分布情况,依次命名为ZmRAV1~ZmRAV4、ZmARF1~ZmARF40、ZmREM1~ZmREM37、ZmHSI1~ZmHSI4和ZmABI1~ZmABI3。各基因的CDS序列长度介于318 (ZmARF25)~3 459 (ZmARF10) bp,编码的肽链由105~1 152个氨基酸组成,分子量介于11.92~127.82 kDa,平均60.10 kDa,等电点(pI)介于5.41~11.31,平均8.00,多数偏碱性。除了ZmARF3具有信号肽无跨膜结构外,其余成员没有信号肽且不存在跨膜结构。亚细胞定位预测发现,大部分家族成员定位于细胞核(63个)上,其余成员分别定位于叶绿体(14个)、细胞质(10个)和细胞骨架(1个)上。
表 1 玉米B3基因家族信息Table 1. Family information associated with B3 gene in maize基因名称
Gene
name基因ID
Gene ID染色体
Chromosome位置
Location蛋白长度
Amino acid
length/aa氨基酸分子质量
Protein molecular
weight/kDa等电点
Isoelectric
point信号肽
Signal
peptide亚细胞定位
Subcellular
localizationZmABI1 Zm00001eb143690 Chr.3 164992981-164997947 691 73.25 6.25 No Nuclear ZmABI2 Zm00001eb259870 Chr.6 5531130-5534996 317 35.05 7.09 No Chloroplast ZmABI3 Zm00001eB361390 Chr.8 158727000-158731148 392 42.64 10.14 No Chloroplast ZmARF1 Zm00001eb029770 Chr.1 162618592-162620659 446 48.73 8.66 No Cytoplasmic ZmARF10 Zm00001eb101720 Chr.2 194575898-194576928 105 11.92 11.02 No Cytoplasmic ZmARF11 Zm00001eb118970 Chr.3 1675067-1679757 684 76.59 6.29 No Nuclear ZmARF12 Zm00001eb133000 Chr.3 94801561-94808087 830 91.69 6.70 No Nuclear ZmARF13 Zm00001eb136130 Chr.3 125302568-125308760 888 97.84 6.08 No Nuclear ZmARF14 Zm00001eb142540 Chr.3 158783741-158788605 805 89.80 6.25 No Nuclear ZmARF15 Zm00001eb152940 Chr.3 201732015-201737265 720 78.34 7.17 No Nuclear ZmARF16 Zm00001eb157270 Chr.3 215856048-215864377 622 68.16 6.93 No Nuclear ZmARF17 Zm00001eb170540 Chr.4 25420464-25426332 811 89.96 7.33 No Nuclear ZmARF18 Zm00001eb182260 Chr.4 123531034-123536559 465 51.97 6.89 No Nuclear ZmARF19 Zm00001eb185290 Chr.4 148952420-148955210 748 79.75 11.10 No Nuclear ZmARF2 Zm00001eb031700 Chr.1 177503771-177511926 897 98.42 6.23 No Nuclear ZmARF20 Zm00001eb202300 Chr.4 220668443-220669476 106 12.10 11.31 No Cytoplasmic ZmARF21 Zm00001eb207450 Chr.4 245046366-245052434 847 93.91 6.16 No Nuclear ZmARF22 Zm00001eb221620 Chr.5 29000492-29005177 644 70.96 6.97 No Nuclear ZmARF23 Zm00001eb224680 Chr.5 43789840-43796267 913 100.91 6.47 No Nuclear ZmARF24 Zm00001eb225570 Chr.5 50935637-50939357 725 78.46 7.04 No Nuclear ZmARF25 Zm00001eb227100 Chr.5 59295296-59302607 1 152 127.82 6.72 No Nuclear ZmARF26 Zm00001eb232120 Chr.5 80470636-80479100 1 147 127.24 6.08 No Chloroplast ZmARF27 Zm00001eb243930 Chr.5 177184418-177190185 673 74.66 6.39 No Nuclear ZmARF28 Zm00001eb247720 Chr.5 190712368-190715567 541 57.43 6.73 No Cytoplasmic ZmARF29 Zm00001eb273540 Chr.6 98831655-98835876 700 75.51 7.56 No Nuclear ZmARF3 Zm00001eb035640 Chr.1 194732533-194747737 1 039 115.70 6.74 Yes Nuclear ZmARF30 Zm00001eb273840 Chr.6 101005067-101012686 665 73.65 6.51 No Nuclear ZmARF31 Zm00001eb292830 Chr.6 170115679-170122390 582 64.12 7.46 No Nuclear ZmARF32 Zm00001eb295830 Chr.6 175975687-175981396 659 71.99 7.77 No Nuclear ZmARF33 Zm00001eB363810 Chr.8 166328792-166334593 812 90.88 6.57 No Nuclear ZmARF34 Zm00001eB370810 Chr.8 180707713-180714633 571 62.70 8.21 No Nuclear ZmARF35 Zm00001eB373970 Chr.9 12448248-12460296 957 106.56 6.86 No Nuclear ZmARF36 Zm00001eb408800 Chr.10 13673999-13679982 813 89.70 7.25 No Nuclear ZmARF37 Zm00001eb426740 Chr.10 133561653-133564154 588 63.90 8.77 No Nuclear ZmARF38 Zm00001eb433020 Chr.10 149374022-149380403 945 103.84 6.32 No Nuclear ZmARF39 Zm00001eb433460 Chr.10 149976350-149985703 748 83.71 6.42 No Nuclear ZmARF4 Zm00001eb045640 Chr.1 236387537-236391953 681 74.54 7.92 No Nuclear ZmARF40 Zm00001eb434380 Chr.10 151402521-151404374 462 50.56 5.41 No Cytoskeleton ZmARF5 Zm00001eb066640 Chr.2 2524918-2533246 752 83.91 6.23 No Nuclear ZmARF6 Zm00001eb067270 Chr.2 3630367-3636867 930 102.17 6.19 No Nuclear ZmARF7 Zm00001eb076420 Chr.2 26933847-26940106 513 55.45 6.4 No0 Cytoplasmic ZmARF8 Zm00001eb077170 Chr.2 29481074-29481902 255 27.31 8.00 No Chloroplast ZmARF9 Zm00001eb082150 Chr.2 49388328-49393787 661 73.29 6.14 No Nuclear ZmHSI1 Zm00001eb106940 Chr.2 211825956-211838415 955 105.22 8.41 No Chloroplast ZmHSI2 Zm00001eb111570 Chr.2 223781937-223797411 793 87.23 5.84 No Nuclear ZmHSI3 Zm00001eB324300 Chr.7 166725324-166733527 957 105.24 8.42 No Chloroplast ZmHSI4 Zm00001eB330690 Chr.7 182217234-182223329 963 105.14 6.80 No Chloroplast ZmRAV1 Zm00001eb156040 Chr.3 212115620-212117266 389 40.49 9.92 No Cytoplasmic ZmRAV2 Zm00001eb294830 Chr.6 174089156-174090850 406 42.89 10.32 No Chloroplast ZmRAV3 Zm00001eB342850 Chr.8 65961433-65962833 375 40.28 10.48 No Cytoplasmic ZmRAV4 Zm00001eB360750 Chr.8 156425137-156426706 405 41.97 9.62 No Chloroplast ZmREM1 Zm00001eb001550 Chr.1 4613829-4616572 327 36.03 10.02 No Nuclear ZmREM10 Zm00001eb051370 Chr.1 260998593-261002924 809 90.56 8.92 No Nuclear ZmREM11 Zm00001eb073010 Chr.2 15684088-15686128 409 43.86 6.13 No Nuclear ZmREM12 Zm00001eb088080 Chr.2 106251779-106258244 542 60.72 10.42 No Nuclear ZmREM13 Zm00001eb124740 Chr.3 19549253-19550940 378 40.33 10.07 No Nuclear ZmREM14 Zm00001eb144270 Chr.3 170522446-170526238 693 76 549.20 6.13 No Nuclear ZmREM15 Zm00001eb161000 Chr.3 227546140-227548914 244 27.69 6.88 No Nuclear ZmREM16 Zm00001eb166640 Chr.4 6162714-6163412 232 26.32 6.75 No Cytoplasmic ZmREM17 Zm00001eb168870 Chr.4 16067079-16070427 503 55.39 10.13 No Chloroplast ZmREM18 Zm00001eb171080 Chr.4 28433030-28440841 232 26.66 8.93 No Nuclear ZmREM19 Zm00001eb187370 Chr.4 161654747-161662833 420 45.23 9.69 No Nuclear ZmREM2 Zm00001eb006360 Chr.1 17899920-17901860 239 27.08 9.50 No Nuclear ZmREM20 Zm00001eb197370 Chr.4 195899273-195901016 307 34.54 7.24 No Nuclear ZmREM21 Zm00001eb218990 Chr.5 19018569-19021152 333 37.50 10.12 No Nuclear ZmREM22 Zm00001eb219020 Chr.5 19067468-19070743 522 57.61 7.29 No Nuclear ZmREM23 Zm00001eb250760 Chr.5 204583184-204586327 422 44.97 7.93 No Nuclear ZmREM24 Zm00001eb268680 Chr.6 65750968-65751648 226 25.47 10.33 No Cytoplasmic ZmREM25 Zm00001eb290900 Chr.6 166115906-166120702 278 31.47 9.63 No Nuclear ZmREM26 Zm00001eB347830 Chr.8 98423808-98424587 259 29.34 9.80 No Nuclear ZmREM27 Zm00001eB361750 Chr.8 159865221-159870386 708 80.49 8.41 No Nuclear ZmREM28 Zm00001eB393690 Chr.9 131280885-131282501 277 29.87 10.42 No Nuclear ZmREM29 Zm00001eb401610 Chr.9 156441390-156442035 169 19.09 6.78 No Nuclear ZmREM3 Zm00001eb022070 Chr.1 84498120-84499710 273 29.65 10.89 No Nuclear ZmREM30 Zm00001eb406950 Chr.10 5668101-5669451 283 31.02 8.03 No Nuclear ZmREM31 Zm00001eb412120 Chr.10 44918969-44919649 226 25.53 9.97 No Nuclear ZmREM32 Zm00001eb413890 Chr.10 66245954-66259521 259 29.58 9.21 No Cytoplasmic ZmREM33 Zm00001eb414160 Chr.10 68351628-68352485 285 31.82 8.64 No Chloroplast ZmREM34 Zm00001eb415750 Chr.10 79426434-79427535 249 27.13 6.72 No Nuclear ZmREM35 Zm00001eb419760 Chr.10 102880836-102884729 435 48.58 10.48 No Chloroplast ZmREM36 Zm00001eb421100 Chr.10 111761405-111762472 159 17.32 8.78 No Chloroplast ZmREM37 Zm00001eb427970 Chr.10 137229874-137233357 404 43.36 7.09 No Nuclear ZmREM4 Zm00001eb030510 Chr.1 168954998-168957929 356 40.69 9.04 No Nuclear ZmREM5 Zm00001eb033180 Chr.1 184762763-184764162 291 30.60 7.64 No Chloroplast ZmREM6 Zm00001eb051330 Chr.1 260844051-260846164 324 36.87 8.66 No Nuclear ZmREM7 Zm00001eb051340 Chr.1 260876137-260878512 337 38.84 10.57 No Nuclear ZmREM8 Zm00001eb051350 Chr.1 260883320-260886825 332 38.01 10.44 No Nuclear ZmREM9 Zm00001eb051360 Chr.1 260939215-260942030 504 57.08 6.50 No Nuclear Nuclear:细胞核;Chloroplast:叶绿体;Cytoplasmic:细胞质;Cytoskeleton:细胞骨架。 2.2 玉米B3基因家族种内聚类及基因结构分析
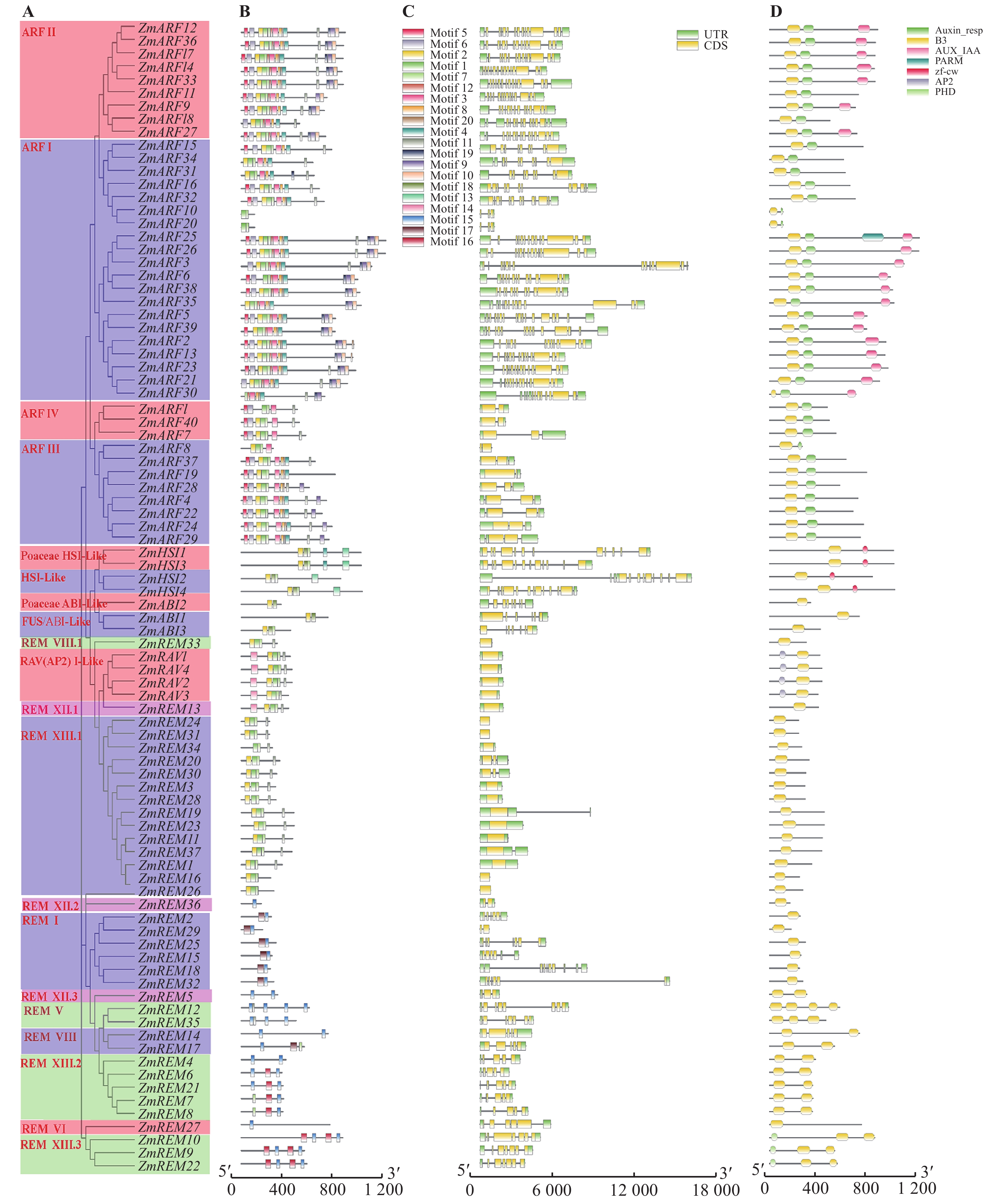
依据玉米B3家族成员的种内聚类及基因结构信息,绘制玉米B3基因家族结构图(图1)。88个玉米B3蛋白中共预测到16个基序,不同成员间不存在完全保守的基序,但聚类在同一分支的成员存在一定的相似保守结构。其中,Motif 1为ZmARF亚家族最保守的基序,40个成员均包含这个基序,其次是Motif 7,除ZmARF30外其他39个成员均含有该基序;而Motif 2和Motif 18是ZmHSI和ZmABI亚家族最保守的基序,分布在这两个亚家族所有成员的氨基酸序列中;另外,Motif 15、Motif 16和Motif 17是ZmREM亚族中特有的基序,且Motif 17是ZmREM36和ZmREM37上唯一含有的基序;ZmRAV亚家族包含的基序极度保守,所有成员都包含Motif 1、Motif 2、Motif 7、Motif 11和Motif 14这5种相同的基序且排列方式一致;ZmB3家族不同亚家族成员之间及同家族内保守基序分布存在差异,暗示玉米中的B3家族成员的功能多样化。
![]() 图 1 玉米B3基因家族的种内聚类、保守基序、基因结构和结构域组成分析A. B3基因家族种内聚类(不同颜色方块代表不同种内聚类);B. B3基因家族保守基序组成与分布(不同颜色方块代表不同保守基序区域,黑色线条代表无保守序列区域);C. B3基因结构组成(绿色区域代表UTR区,黄色区域表示CDS区,黑色线条表示内含子);D. B3基因家族保守结构域(不同颜色方块代表不同保守结构域,黑色线条代表无保守结构区域)。Figure 1. Analysis of intraspecies clustering, conserved motifs, gene structure, and domain composition of B3 gene family in maizeA. Intraspecific clustering of B3 gene family (different colored squares represent different intraspecific clustering); B. Composition and distribution of conservative motifs in the B3 gene family (different colored squares represent different regions of conservative motifs, and black lines represent regions without conserved sequence); C. B3 gene structure composition (green area represents UTR region, yellow area represents CDS region, and black line represents intron); D. Conservative domains of B3 gene family (different colored squares represent different conservative domains, and black lines represent regions without conservative structures).
图 1 玉米B3基因家族的种内聚类、保守基序、基因结构和结构域组成分析A. B3基因家族种内聚类(不同颜色方块代表不同种内聚类);B. B3基因家族保守基序组成与分布(不同颜色方块代表不同保守基序区域,黑色线条代表无保守序列区域);C. B3基因结构组成(绿色区域代表UTR区,黄色区域表示CDS区,黑色线条表示内含子);D. B3基因家族保守结构域(不同颜色方块代表不同保守结构域,黑色线条代表无保守结构区域)。Figure 1. Analysis of intraspecies clustering, conserved motifs, gene structure, and domain composition of B3 gene family in maizeA. Intraspecific clustering of B3 gene family (different colored squares represent different intraspecific clustering); B. Composition and distribution of conservative motifs in the B3 gene family (different colored squares represent different regions of conservative motifs, and black lines represent regions without conserved sequence); C. B3 gene structure composition (green area represents UTR region, yellow area represents CDS region, and black line represents intron); D. Conservative domains of B3 gene family (different colored squares represent different conservative domains, and black lines represent regions without conservative structures).在基因结构方面,处于不同聚类分支下的家族成员外显子个数差距丰富,可具备单个或多个外显子,但同支间外显子个数相对保守,其中RAVI-Like亚组成员只包含1个外显子的典型结构,而ARF II亚族成员的外显子数介于13~14个,含有的外显子数目最多。玉米B3不同亚族成员外显子分布展现的多样性和相似性,进一步表明其基因结构在进化中可能发生了分化并进一步影响功能的变异。
结构域组成分析结果显示,玉米B3家族成员除含B3结构域外,部分成员还包括Auxin_resp、AUX_IAA、PHD、zf-CW、AP2、PARM等结构域。其中ABI亚族成员仅包含B3结构域,说明该亚族在植物进化过程中高度保守;ARF亚族成员含有的结构域最为丰富,此亚族基因编码的蛋白含有B3、Auxin_resp、AUX_IAA、PARM 4种结构域,可能是ARF亚族在进化过程中发生了一些适应性的变异。
2.3 玉米及拟南芥、水稻B3基因家族系统发育树的构建
将分别来自玉米、拟南芥和水稻的88、87和85个B3基因,依据编码的氨基酸序列保守结构分别划分为5个亚家族,并采用NJ法(重复1 000次)构建不同亚族间的系统进化树(图2)。拟南芥与水稻中B3家族成员均分别聚类在玉米B3家族的5个亚族中,且每个亚族可分为许多小分支。例如ABI亚族可分为3个分支,ZmABI1、ZmABI2以及ZmABI3分别属于这3个分支;HSI亚族可分为两个分支,ZmHSI1和ZmHSI3亲缘关系较近,属于同一个分支,而ZmHSI2和ZmHSI4属于另一分支;RAV亚族可分为两个分支,ZmRAV1、ZmRAV4在一个分支内,ZmRAV2与ZmRAV3则在另一分支内;ARF亚族可分为4个分支,ARF Ⅳ分支的成员较少,仅含有ZmARF1、ZmARF7、ZmARF40基因;REM亚族可分为4个分支,ZmREM27和ZmREM36分别单独聚为一类,与同族其他基因亲缘关系较远。系统进化分析的结果表明:玉米B3家族与水稻B3家族成员间的亲缘关系更为密切,可能具有相似的基因功能。
2.4 玉米B3家族基因的顺式作用元件分析
对玉米B3基因的启动子区进行顺式作用元件预测,发现其启动子区域包含多种响应激素和逆境胁迫的作用元件(图3)。首先,玉米B3家族成员的启动子区中至少存在1个激素响应元件,70个成员含有脱落酸响应元件(ABRE);34个成员含有生长素响应元件(TGA-box);73个成员含有茉莉酸甲酯响应元件(TGACG-motif);30个成员含有赤霉素响应元件(GARE-motif);11个成员含有水杨酸响应元件(TCA-element)。ZmREM31含有数量最丰富的激素响应元件(15个)。其次在胁迫响应元件的分析中发现,63个成员含有厌氧诱导响应元件(GC-motif);26个成员含有低温响应元件(LTR);30个成员含有干旱诱导响应元件(MBS);16个成员含有防御和压力应答元件(TC-rich repeats)。ZmREM17所含胁迫响应元件数量最多(9个),其中7个属于低温响应元件(LTR),推测其在低温胁迫应答中扮演着重要的角色。另外,还在部分成员启动子区域中鉴定到响应响应调控胚乳表达(GCN4-motif)、种子萌发(RY-element)、细胞周期(MSA-like)、分生组织表达(CAT-box)、昼夜节律(Circadian)等与发育相关的作用元件。以上结果表明玉米B3家族成员在环境胁迫响应、激素合成转导和玉米生长发育中参与应激反应及调控,并存在多种应答方式。
2.5 玉米B3基因家族的组织特异性表达分析
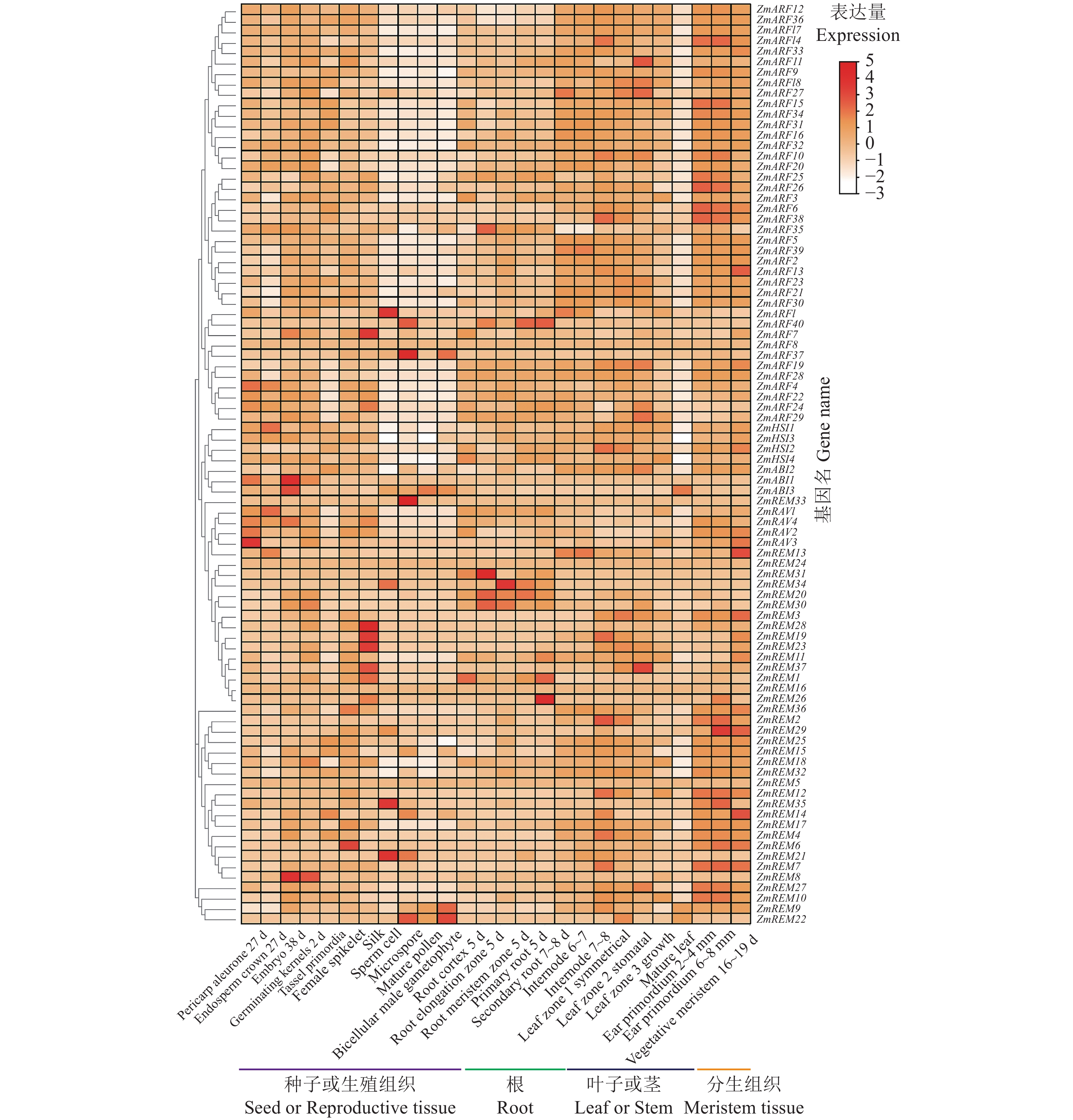
本研究中,TPM值用于表示B3家族基因在不同玉米组织中的相对表达水平,利用Tbtools软件将组织特异性表达进行可视化(图4)。结果表明,玉米B3基因家族成员在不同部位的表达情况不同:在2~4节的节间组织中,ZmARF26、ZmREM27等基因优势表达;在种子、花器官等生殖器官中,ZmREM22、ZmABI3、ZmARF1、ZmARF37等基因表达量较高;而在叶或及茎等上端营养组织中,ZmHSI3、ZmARF15、ZmARF16、ZmARF18、ZmARF24、ZmARF29等均产生较高水平的表达;对于分生组织来说,大部分的ARF及REM亚族基因、ZmRAV2、ZmRAV3等均发生表达上调。其中,同处于一类的组织,如根的不同部位也存在选择性表达,例如根冠部分和主根、次级根分别为ZmREM1 + ZmREM31 + ZmHSI4、ZmREM34 + ZmREM20 + ZmARF40、ZmREM26 + ZmREM1 + ZmREM11 + ZmARF40等几个基因表达较强,且次生根中基因的表达量最高。值得注意的是,ZmREM5、ZmREM24和ZmARF8在所有组织中几乎都不表达。这些结果表明,玉米B3家族基因在不同的组织器官中有差异表达,并参与了玉米生长发育的不同阶段。
![]() 图 4 玉米B3基因家族的组织特异性表达分析Pericarp aleurone 27 d:授粉后27 d果皮/糊粉层;Endosperm crown 27 d:授粉后27 d胚乳冠;Embryo 38 d:授粉后38 d胚胎;Germinating kernels 2 d:籽粒发芽后2 d;Tassel primordia:穗原基;Female spikelet:雌性小穗;Silk:穗丝;Sperm cell:精细胞;Microspore:花粉粒;Mature pollen:成熟花粉;Bicellular male gametophyte:双核雄性配子体;Root cortex 5 d:根皮层5 d;Root elongation zone:根-伸长区5 d;Root meristem zone 5 d:根-分生组织区5 d;Primary root 5 d:主根5 d;Secondary root 7~8 d:次生根7~8 d;Internode 6~7:6~7节间;Internode 7~8:7~8节间;Leaf zone 1 symmetrical:叶区1 (对称);Leaf zone 2 stomatal:叶区2 (气孔);Leaf zone 3 growth:叶区3 (生长);Mature leaf:成熟叶;Ear primordium 2~4 mm:耳原基2~4 mm;Ear primordium 6~8 mm:耳原基6~8 mm;Vegetative meristem 16~19 d:植物分生组织16~19 d。Figure 4. Tissue-specific expression analysis of B3 gene family in maizePericarp aleurone 27 d: pericarp/aleurone layer 27 d after pollination; Endosperm crown 27 d: endosperm crown 27 d after pollination; Embryo 38 d: embryos 38 d after pollination; Germinating kernels 2 d: seeds 2 d after germination; Tassel primordia: spike primordium; Female spikelet: female spikelet; Silk: ear filaments; Sperm cell: spermatid; Microspore: pollen grains; Mature pollen: mature pollen; Bicellular male gametophyte: binuclear male gametophyte; Root cortex 5 d: root cortex 5 d; Root elongation zone: root elongation zone 5 d; Root meristem zone 5 d: root meristem zone 5 d; Primary root 5 d: main root 5 d; Secondary root 7~8 d: secondary root 7~8 d; Internode 6~7: 6~7 internode; Internode 7~8: 7~8 internode; Leaf zone 1 symmetrical: leaf zone 1 (symmetrical); Leaf zone 2 stomal: Leaf area 2 (stomata); Leaf zone 3 growth: leaf zone 3 (growth); Mature leaf: mature leaf; Ear primordium 2~4 mm: ear primordium 2~4 mm; Ear primordium 6~8 mm: ear primordium 6~8 mm; Vegetative meristem 16~19 d: plant meristem 16~19 d.
图 4 玉米B3基因家族的组织特异性表达分析Pericarp aleurone 27 d:授粉后27 d果皮/糊粉层;Endosperm crown 27 d:授粉后27 d胚乳冠;Embryo 38 d:授粉后38 d胚胎;Germinating kernels 2 d:籽粒发芽后2 d;Tassel primordia:穗原基;Female spikelet:雌性小穗;Silk:穗丝;Sperm cell:精细胞;Microspore:花粉粒;Mature pollen:成熟花粉;Bicellular male gametophyte:双核雄性配子体;Root cortex 5 d:根皮层5 d;Root elongation zone:根-伸长区5 d;Root meristem zone 5 d:根-分生组织区5 d;Primary root 5 d:主根5 d;Secondary root 7~8 d:次生根7~8 d;Internode 6~7:6~7节间;Internode 7~8:7~8节间;Leaf zone 1 symmetrical:叶区1 (对称);Leaf zone 2 stomatal:叶区2 (气孔);Leaf zone 3 growth:叶区3 (生长);Mature leaf:成熟叶;Ear primordium 2~4 mm:耳原基2~4 mm;Ear primordium 6~8 mm:耳原基6~8 mm;Vegetative meristem 16~19 d:植物分生组织16~19 d。Figure 4. Tissue-specific expression analysis of B3 gene family in maizePericarp aleurone 27 d: pericarp/aleurone layer 27 d after pollination; Endosperm crown 27 d: endosperm crown 27 d after pollination; Embryo 38 d: embryos 38 d after pollination; Germinating kernels 2 d: seeds 2 d after germination; Tassel primordia: spike primordium; Female spikelet: female spikelet; Silk: ear filaments; Sperm cell: spermatid; Microspore: pollen grains; Mature pollen: mature pollen; Bicellular male gametophyte: binuclear male gametophyte; Root cortex 5 d: root cortex 5 d; Root elongation zone: root elongation zone 5 d; Root meristem zone 5 d: root meristem zone 5 d; Primary root 5 d: main root 5 d; Secondary root 7~8 d: secondary root 7~8 d; Internode 6~7: 6~7 internode; Internode 7~8: 7~8 internode; Leaf zone 1 symmetrical: leaf zone 1 (symmetrical); Leaf zone 2 stomal: Leaf area 2 (stomata); Leaf zone 3 growth: leaf zone 3 (growth); Mature leaf: mature leaf; Ear primordium 2~4 mm: ear primordium 2~4 mm; Ear primordium 6~8 mm: ear primordium 6~8 mm; Vegetative meristem 16~19 d: plant meristem 16~19 d.2.6 玉米B3基因家族的胁迫应答分析
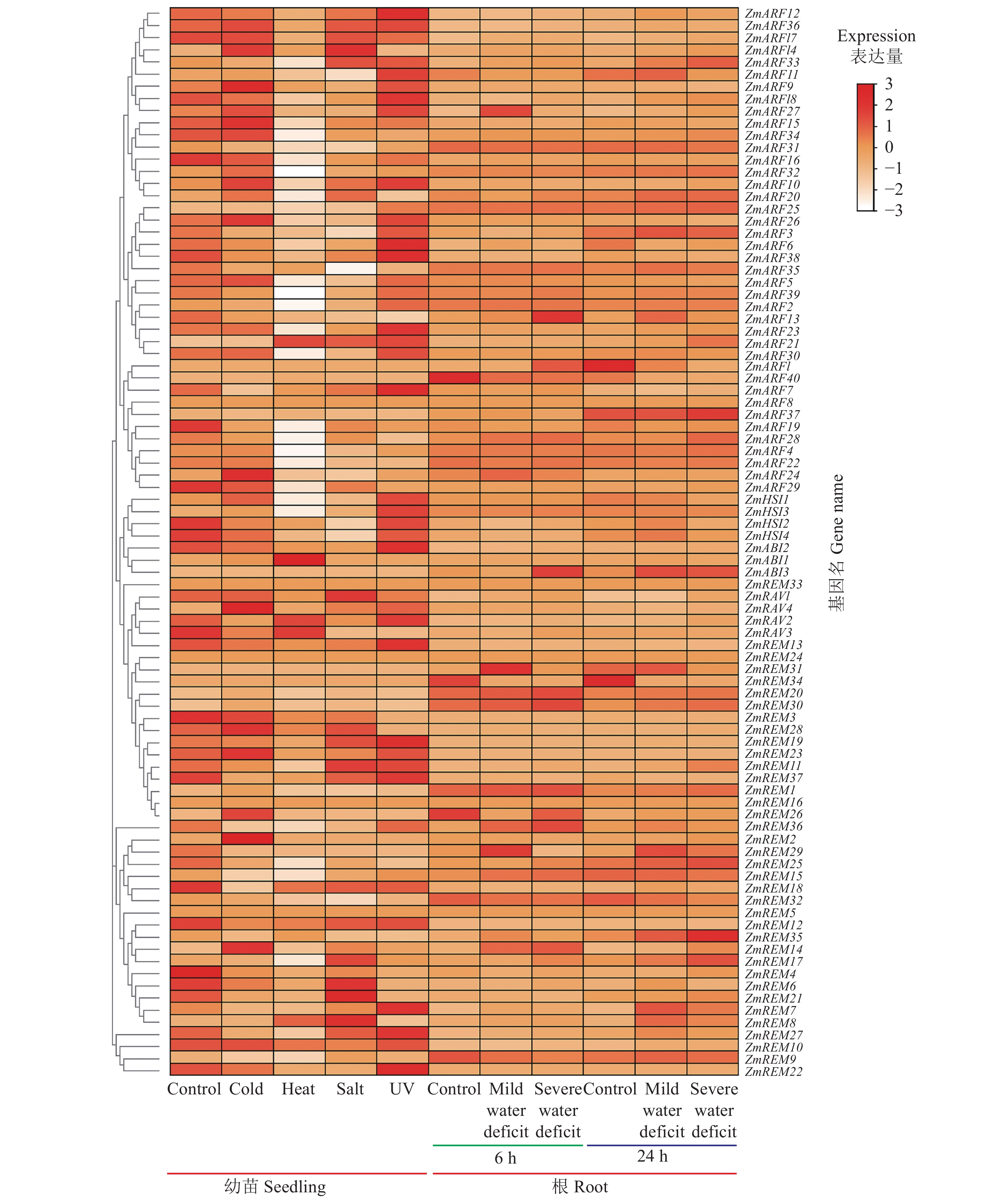
为解析玉米B3基因家族成员对胁迫因子的响应,进一步挖掘所有玉米B3基因在冷、热、盐、紫外和干旱5种逆境胁迫下的表达模式,结果显示大部分的玉米B3基因家族成员均可对非生物胁迫因子产生应激表现(图5)。不同玉米B3基因对不同逆境胁迫表现出不同程度的表达水平调节,例如冷胁迫处理后,46个基因下调表达,32个基因上调表达,ZmREM2、ZmRAV4基因在处理后表达上调最为显著,分别上调4.8和5倍;热胁迫处理后,53个基因表达下调,9个基因表达上调,ZmREM30、ZmABI1对热胁迫响应剧烈,热处理后是处理前的3.3和4.8倍;盐胁迫下表达上调的20个基因中,ZmARF14、 ZmRAV4、ZmREM30、ZmREM8等基因对盐胁迫响应强烈,其表达量是对照的2.2、2.3、6.9、2.8倍,暗示ZmREM30在抵御盐胁迫通路中发挥重要作用;而ZmREM22、ZmREM7、ZmREM13、ZmHSI1~4、ZmARF6、ZmARF7等40个基因可响应紫外线处理导致的氧化胁迫反应,ZmHSI3对紫外胁迫响应最强烈,表达上调1.7倍。而在叶片中,也显示出多样的非生物逆境响应特征,同时具备一定的响应时间选择特异性,如ZmREM30、ZmREM20、ZmARF13、ZmABI3可对6 h的严重缺水胁迫产生响应,而严重缺水胁迫持续到24 h时,表达出现下调,这些基因可能在6 h的严重缺水胁迫发挥关键作用,ZmREM25、ZmREM35、ZmARF37则在24 h的严重缺水胁迫发挥主要调控作用,ZmREM35在严重缺水处理24 h后,表达量上升到对照处理的1.7倍。玉米B3基因家族成员对在胁迫环境下不同程度的诱导表达,与其启动子中存在丰富的逆境胁迫响应元件呼应吻合,佐证了玉米B3基因家族在逆境抗性中的重要作用。
![]() 图 5 玉米B3基因家族的胁迫应答分析Control:对照;Cold:冷胁迫;Heat:热胁迫;Salt:盐胁迫;UV:紫外线胁迫;Mild water deficit:轻度干旱胁迫;Severe water deficit:重度干旱胁迫。Figure 5. Stress response analysis of B3 gene family in maizeCold: cold stress; Heat: heat stress; Salt: salt stress; UV: UV stress; Mild water deficit: mild drought stress: severe water deficit: severe drought stress.
图 5 玉米B3基因家族的胁迫应答分析Control:对照;Cold:冷胁迫;Heat:热胁迫;Salt:盐胁迫;UV:紫外线胁迫;Mild water deficit:轻度干旱胁迫;Severe water deficit:重度干旱胁迫。Figure 5. Stress response analysis of B3 gene family in maizeCold: cold stress; Heat: heat stress; Salt: salt stress; UV: UV stress; Mild water deficit: mild drought stress: severe water deficit: severe drought stress.3. 讨论
本研究运用生物信息学手段对玉米中B3家族基因进行鉴定,共获得88个玉米B3家族成员,略多于Wang等[4] 81个玉米B3家族成员的鉴定结果,推测可能因参照基因组版本及筛选阈值不同所导致。其所含氨基酸数量介于105~1 052,而陆地棉(Gossypium hirsutum)的79个B3氨基酸长度介于119~749 [30],番茄(Solanum lycopersicum)的97个B3氨基酸长度介于92~1 317 [31],表示B3蛋白氨基酸序列长度在植物中相对保守。根据亚细胞定位预测,63个基因定位在细胞核,其余成员分别定位在叶绿体、细胞质和细胞骨架上。除特有的B3结构域外,一些成员还包括Auxin_resp、AUX_IAA、PHD、zf-CW、AP2、PARM等结构域,依据结构域和进化关系,将玉米B3家族分为ARF、ABI、HSI、RAV和REM 5个亚家族,这5个亚族所包含的B3基因数目差别很大,其中,ARF (40个)和REM (37个)亚族成员数目最多,RAV (4个)、HSI3 (4个)亚族成员较少,ABI (3个)亚族成员最少。Wang等[4]在拟南芥、高粱(Sorghum bicolor)和水稻中鉴定到ABI家族数量分别为3、4、5个,数量相比B3家族其他的基因少许多,与本研究的结果一致。同一亚家族的成员结构相似,可以较好聚为一类,但家族间的差异较大,体现出相同亚族成员结构的同源性及稳定性。分析保守基序发现大多数ZmB3s家族成员都含有Motif1和Motif7,这两种基序保守性较高。与之相反,一些基序仅存在于特定的亚家族中,例如,Motif13只存在于ZmHSI亚家族,Motif15、Motif16和Motif17仅存在于ZmREM亚家族,石荣康等[30]在陆地棉REM基因家族的研究中也报道了基序分布的特异性,推测这些基序可能在玉米进化中发挥特定功能。预测和分析启动子区的顺式作用元件可以推测下游基因所编码蛋白质的可能功能。顺式元件分析结果显示,88个ZmB3s基因上游启动子区分布的顺式元件类型与数量不同,这可能是导致其基因功能发生分化的主要原因,大部分ZmB3s基因富含激素和胁响应元件,这表明B3家族成员可能通过参与不同的信号转导途径,在植物生长发育和抗逆胁迫中发挥作用,其中茉莉酸甲酯响应元件(TGACG-motif)最为丰富,暗示ZmB3家族成员可能在茉莉酸甲酯信号通路中发挥重要调节作用。
本研究发现,ZmB3s基因的表达具有组织特异性。ZmARF家族(如ZmARF5)在结节和叶中的表达量较高,与甘草(Glycyrrhiza uralensis) GuARF5在茎叶中高表达并参与植物生长结果一致[32],推测ZmARF家族在参与玉米快速生长中发挥作用。ZmHSI1、ZmRAV4在胚乳和分生组织中表达量上调,刘颖慧和董志平[5]发现拟南芥中AtHSI2调控糖介导的信号响应,促进种子的萌发;李庆飞等[33]发现CmRAV在叶片中表达水平最高,参与南瓜(Cucurbita moschata)生育器官雌雄花和营养器官叶片的发育。ZmREM家族在不同组织间存在差异表达,ZmREM29在分生组织表达量较高,ZmREM31、ZmREM34在根中表达量变化显著,这与瓮巧云等[34]报道的ZmREM基因在根中表达水平较高不同,推测同家族的REM基因因结构的差异也可能引起组织表达模式变化。而ZmREM9、ZmREM32、ZmARF39等14个基因在所有组织中均有较高表达水平,暗示这些基因可能在玉米整个生命周期中都发挥调控作用。
对B3家族逆境胁迫下的转录组分析发现,大部分B3基因对冷、热、盐、紫外胁迫中至少一种响应强烈。本研究中ZmREM2和ZmRAV4在响应低温胁迫时表达上调,ZmARF14、ZmRAV4、ZmREM30在响应盐胁迫时表达显著上调,ZmABI3、ZmREM20、ZmARF37响应干旱胁迫,并表现出时间选择的特异性。有报道发现,过表达GmRAV可以提高南瓜株系对高盐和干旱的抗性[35],玉米ZmREM受干旱、盐、冷和热胁迫等的诱导表达[34],ZmRAV1的表达受到脱水、高盐和ABA等胁迫的诱导,与野生型植株相比,ZmRAV1过表达转基因株系提高了对盐胁迫和渗透胁迫的抗性[36],推测ZmRAV4、ZmREM20等相关基因可能具有一因多效的功能。此外,ZmREM22、ZmREM7、ZmHSI3在紫外胁迫时显著诱导表达。ABI亚族中,ZmABI1在响应高温胁迫时表达量显著变化,ZmABI3受干旱胁迫诱导表达,这与Weng 等[37]发现ZmABI的表达受多种非生物胁迫(包括高温、高盐、低温、干旱)的诱导结果一致。
4. 结论
本研究从玉米基因组鉴定出88个B3基因家族成员,确定了基因在染色体上的位置,分析了基因结构、理化性质以及系统发育特征,通过玉米芯片数据和转录组数据分析了B3家族成员在生长发育和逆境胁迫响应中的功能,相关研究结果将为B3家族基因的功能研究及调控玉米的发育研究提供理论参考。
参考文献
[1] SASNAUSKAS G, MANAKOVA E, LAPĖNAS K, KAUNECKAITĖ K, SIKSNYS V. DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. Febs Journal, 2018, 285(21): 4041-4059. doi: 10.1111/febs.14649
[2] WANG W B, AO T, ZHANG Y Y, WU D, XU W, HAN B, LIU A Z. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis L.). Plant Diversity, 2021, 44(2): 201-212.
[3] BARRETO H G, SÁGIO S A, JÚNIOR A C, FEVEREIRO P, BENEDITO V A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. Plant Cell Tissue and Organ Culture, 2019, 139(2): 327-337. doi: 10.1007/s11240-019-01687-3
[4] WANG Y, DENG D, ZHANG R, WANG S, BIAN Y, YIN Z. Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Molecular Biology Reports, 2012, 39(5): 6267-6282. doi: 10.1007/s11033-012-1448-8
[5] 刘颖慧, 董志平. 植物B3转录因子的结构和功能. 分子植物育种, 2017, 15(5): 1868-1873. LIU Y H, DONG Z P. Structure and function of plant B3 transcription factor. Molecular Plant Breeding, 2017, 15(5): 1868-1873.
[6] ROMANEL E A, SCHRAGO C G, COUÑAGO R M, RUSSO C A, ALVES-FERREIRA M. Evolution of the B3 DNA binding superfamily, new insights into rem family gene diversification. PLoS One, 2009, 4(6): 1-18.
[7] 吴佳军, 俞率成, 刘志刚, 傅鹰, 周明兵. 毛竹B3家族全基因组鉴定及表达模式分析. 农业生物技术学报, 2019, 27(1): 43-54. WU J J, YU S C, LIU Z G, FU Y, ZHOU M B. Genome wide identification and expression pattern analysis of pubescens B3 family in Phyllostachys. Journal of Agricultural Biotechnology, 2019, 27(1): 43-54.
[8] VERMA S, BHATIA S. A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum L.). 3 Biotech, 2019, 9(9): 1-17.
[9] CHANDLER J W. Auxin response factors. Plant Cell and Environment, 2016, 39(5): 1014-1028. doi: 10.1111/pce.12662
[10] LI S B, XIE Z Z, HU C G, ZHANG J Z. A review of auxin response factors (ARFs) in plants. Frontiers in Plant Science, 2016, 7(1): 47-48.
[11] LIU Z, MIAO L, HUO R, SONG X, JOHNSON C, KONG L, SUNDARESAN V, YU X. ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant and Cell Physiology, 2018, 59(1): 179-189.
[12] WILMOTH J C, WANG S, TIWARI S B, JOSHI A D, HAGEN G, GUILFOYLE T J, ALONSO J M, ECKER J R, REED J W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal, 2005, 43(1): 118-130. doi: 10.1111/j.1365-313X.2005.02432.x
[13] GUO F, ZHANG P, WU Y, LIAN G, YANG Z, LIU W, BUERTE B, ZHOU C, ZHANG W, LI D, HAN N, TONG Z, ZHU M, XU L, CHEN M, BIAN H. Rice LEAFY COTYLEDON1 hinders embryo greening during the seed development. Frontiers in Plant Science, 2022, 13(1): 1-18.
[14] FREITAS N C, BARRETO H G, TORRES L F, FREIRE L L, RODRIGUES L, DINIZ L, BEIJO L A. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica, competence acquisition and developmental marker genes. Plant Cell Tissue and Organ Culture, 2019, 137(3): 599-611. doi: 10.1007/s11240-019-01594-7
[15] ZINSMEISTER J, LEPRINCE O, BUITINK J. Molecular and environmental factors regulating seed longevity. Biochemical Journal, 2020, 477(2): 305-323. doi: 10.1042/BCJ20190165
[16] KHANDELWAL A, CHO S H, MARELLA H, SAKATA Y, PERROUD P F, PAN A, QUATRANO R S. Role of ABA and ABI3 in desiccation tolerance. Science, 2010, 327: 546-547. doi: 10.1126/science.1183672
[17] 张一弓, 张怡, 阿依白合热木·木台力甫, 张道远. 异源过表达齿肋赤藓ScABI3基因改变拟南芥气孔表型并提高抗旱性. 植物学报, 2021, 56(4): 414-421. doi: 10.11983/CBB20212 ZHANG Y G, ZHANG Y, Ayibaiheremu·Mutailifu, ZHANG D Y. Heterologous overexpression of ScABI3 gene in Erythritus denticola alters the stomatal phenotype of Arabidopsis thaliana and improves drought resistance. Acta Botanica Sinica, 2021, 56(4): 414-421. doi: 10.11983/CBB20212
[18] KIM H U, JUNG S J, LEE K R, KIM E H, LEE S M, ROH K H, KIM J B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio, 2014, 4(1): 25-32. doi: 10.1016/j.fob.2013.11.003
[19] VEERAPPAN V, WANG J, KANG M, LEE J, TANG Y, JHA A K, SHI H, PALANIVELU R, ALLEN R D. A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to depression of seed-specific gene expression. Planta, 2012, 236(1): 1-17. doi: 10.1007/s00425-012-1630-1
[20] CHEN N, VEERAPPAN V, ABDELMAGEED H, KANG M, ALLEN RD. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell, 2018, 30(3): 600-619. doi: 10.1105/tpc.17.00655
[21] LI X J, LI M, ZHOU Y, HU S, HU R, CHEN Y, LI X B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One, 2015, 10(2): 1-18.
[22] PENG Z, WANG M, ZHANG L, JIANG Y, ZHAO C, SHAHID M Q, BAI Y, HAO J, PENG J, GAO Y, SU W, YANG X. EjRAV1/2 Delay Flowering Through Transcriptional Repression of EjFTs and EjSOC1s in Loquat. Frontiers in Plant Science, 2021, 12(1): 1-14.
[23] KING G J, CHANSON A H, MCCALLUM E J, OHME-TAKAGI M, BYRIEL K, HILL J M, MARTIN J L, MYLNE J S. The Arabidopsis B3 domain protein VERNALIZATION1 (VRN1) is involved in processes essential for development, with structural and mutational studies revealing its DNA-binding surface. Journal of Biological Chemistry, 2013, 288(5): 3198-3207. doi: 10.1074/jbc.M112.438572
[24] CASELLI F, BERETTA V M, MANTEGAZZA O, PETRELLA R, LEO G, GUAZZOTTI A, HERRERA-UBALDO H, DE FOLTER S, MENDES M A, KATER M M, GREGIS V. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Frontiers in Plant Science, 2019, 10(2): 1351-1368.
[25] WANG W, ZHANG X, DENG F, YUAN R, SHEN F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genomics, 2017, 18(1): 376-401. doi: 10.1186/s12864-017-3768-5
[26] WARMAN C, PANDA K, VEJLUPKOVA Z, HOKIN S, UNGER-WALLACE E, COLE R A, CHETTOOR A M, JIANG D, VOLLBRECHT E, EVANS M M S, SLOTKIN R K, FOWLER J E. High expression in maize pollen correlates with genetic contributions to pollen fitness as well as with coordinated transcription from neighboring transposable elements. PLoS Genetics, 2020, 16(4): 1-32.
[27] WALLEY J W, SARTOR R C, SHEN Z, SCHMITZ R J, WU K J, URICH M A, NERY J R, SMITH L G, SCHNABLE J C, ECKER J R, BRIGGS S P. Integration of omic networks in a developmental atlas of maize. Science, 2016, 353: 814-818. doi: 10.1126/science.aag1125
[28] MAKAREVITCH I, WATERS A J, WEST P T, STITZER M, HIRSCH C N, ROSS-IBARRA J, SPRINGER N M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genetics, 2015, 11(1): 1-12.
[29] OPITZ N, PASCHOLD A, MARCON C, MALIK W A, LANZ C, PIEPHO H P, HOCHHOLDINGER F. Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genomics, 2014, 15(1): 741-754. doi: 10.1186/1471-2164-15-741
[30] 石荣康, 张冬梅, 孙正文, 刘正文, 解美霞, 张艳, 马峙英, 王省芬. 陆地棉REM基因家族全基因组鉴定及表达分析. 棉花学报, 2021, 33(2): 95-111. doi: 10.11963/1002-7807.srkwxf.20210113 SHI R K, ZHANG D M, SUN Z W, LIU Z W, XIE M X, ZHANG Y, MA S Y, WANG S F. Complete genome identification and expression analysis of REM gene family in Gossypium hirsutum. Cotton Science, 2021, 33(2): 95-111. doi: 10.11963/1002-7807.srkwxf.20210113
[31] 孙亭亭, 王大伟, 龚达平, 陈乐, 陈雅琼, 孙玉合. 番茄B3超家族成员鉴定及生物信息学分析. 植物遗传资源学报, 2015, 16(4): 806-814. SUN T T, WANG D W, GONG D P, CHEN L, CHEN Y Q, SUN Y H. Identification and bioinformatics analysis of tomato B3 superfamily members. Journal of Plant Genetic Resources, 2015, 16(4): 806-814.
[32] 关思静, 高静, 徐蓉蓉, 葛甜甜, 王楠, 颜永刚, 张岗, 陈莹, 刘阿萍, 程萌格. 甘草生长素反应因子(ARF)基因家族的鉴定及表达分析. 中国农学通报, 2021, 37(29): 20-27. doi: 10.11924/j.issn.1000-6850.casb2020-0304 GUAN S J, GAO J, XU R R, GE T T, WANG N, YAN Y G, ZHANG G, CHEN Y, LIU A P, CHENG M G. Identification and expression analysis of auxin response factor (ARF) gene family in Glycyrrhiza uralensis. Chinese Agricultural Science Bulletin, 2021, 37(29): 20-27. doi: 10.11924/j.issn.1000-6850.casb2020-0304
[33] 李庆飞, 王梦梦, 郭卫丽, 周俊国, 李新峥. 南瓜转录因子基因CmRAV的克隆与表达分析. 植物生理学报, 2021, 57(12): 2337-2345. doi: 10.13592/j.cnki.ppj.2021.0144 LI Q F, WANG M M, GUO W L, ZHOU J G, LI X Z. Cloning and expression analysis of pumpkin transcription factor gene CmRAV. Chinese Journal of Plant Physiology, 2021, 57(12): 2337-2345. doi: 10.13592/j.cnki.ppj.2021.0144
[34] 瓮巧云, 赵彦敏, 张贺, 宋晋辉, 马海莲, 袁进成, 刘颖慧. 玉米ZmREM基因的克隆与逆境胁迫后表达分析. 浙江农业学报, 2016, 28(11): 1822-1827. doi: 10.3969/j.issn.1004-1524.2016.11.03 WENG Q Y, ZHAO Y M, ZHANG H, SONG J H, MA H L, YUAN J C, LIU Y H. Cloning and expression analysis of ZmREM gene in maize. Acta Agriculturae Zhejiangensis, 2016, 28(11): 1822-1827. doi: 10.3969/j.issn.1004-1524.2016.11.03
[35] ZHAO S P, XU Z S, ZHENG W J, ZHAO W, WANG Y X, YU T F, CHEN M, ZHOU Y B, MIN D H, MA Y Z, CHAI S C, ZHANG X H. Genome-wide analysis of the RAV family in soybean and functional identification of GMRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Frontiers in Plant Science, 2017, 6(8): 905-916.
[36] 闵浩巍. 玉米抗逆相关基因ZmRAV1和ZmTCX8.1的功能研究. 北京: 中国农业大学博士学位论文, 2013. MIN H W. Study on the function of maize stress resistance related genes ZmRAV1 and ZmTCX8.1. PhD Thesis. Beijing: China Agricultural University, 2013.
[37] WENG Q Y, S ONG J H, MA H L, YUAN J C, ZHAO Y M, ZHAO Y, LIU Y H. Cloning and expression analysis of ZmABI3 gene in Zea mays. Turkish Journal of Biochemistry, 2017, 42(3): 279-285.
-
图 1 玉米B3基因家族的种内聚类、保守基序、基因结构和结构域组成分析
A. B3基因家族种内聚类(不同颜色方块代表不同种内聚类);B. B3基因家族保守基序组成与分布(不同颜色方块代表不同保守基序区域,黑色线条代表无保守序列区域);C. B3基因结构组成(绿色区域代表UTR区,黄色区域表示CDS区,黑色线条表示内含子);D. B3基因家族保守结构域(不同颜色方块代表不同保守结构域,黑色线条代表无保守结构区域)。
Figure 1. Analysis of intraspecies clustering, conserved motifs, gene structure, and domain composition of B3 gene family in maize
A. Intraspecific clustering of B3 gene family (different colored squares represent different intraspecific clustering); B. Composition and distribution of conservative motifs in the B3 gene family (different colored squares represent different regions of conservative motifs, and black lines represent regions without conserved sequence); C. B3 gene structure composition (green area represents UTR region, yellow area represents CDS region, and black line represents intron); D. Conservative domains of B3 gene family (different colored squares represent different conservative domains, and black lines represent regions without conservative structures).
图 4 玉米B3基因家族的组织特异性表达分析
Pericarp aleurone 27 d:授粉后27 d果皮/糊粉层;Endosperm crown 27 d:授粉后27 d胚乳冠;Embryo 38 d:授粉后38 d胚胎;Germinating kernels 2 d:籽粒发芽后2 d;Tassel primordia:穗原基;Female spikelet:雌性小穗;Silk:穗丝;Sperm cell:精细胞;Microspore:花粉粒;Mature pollen:成熟花粉;Bicellular male gametophyte:双核雄性配子体;Root cortex 5 d:根皮层5 d;Root elongation zone:根-伸长区5 d;Root meristem zone 5 d:根-分生组织区5 d;Primary root 5 d:主根5 d;Secondary root 7~8 d:次生根7~8 d;Internode 6~7:6~7节间;Internode 7~8:7~8节间;Leaf zone 1 symmetrical:叶区1 (对称);Leaf zone 2 stomatal:叶区2 (气孔);Leaf zone 3 growth:叶区3 (生长);Mature leaf:成熟叶;Ear primordium 2~4 mm:耳原基2~4 mm;Ear primordium 6~8 mm:耳原基6~8 mm;Vegetative meristem 16~19 d:植物分生组织16~19 d。
Figure 4. Tissue-specific expression analysis of B3 gene family in maize
Pericarp aleurone 27 d: pericarp/aleurone layer 27 d after pollination; Endosperm crown 27 d: endosperm crown 27 d after pollination; Embryo 38 d: embryos 38 d after pollination; Germinating kernels 2 d: seeds 2 d after germination; Tassel primordia: spike primordium; Female spikelet: female spikelet; Silk: ear filaments; Sperm cell: spermatid; Microspore: pollen grains; Mature pollen: mature pollen; Bicellular male gametophyte: binuclear male gametophyte; Root cortex 5 d: root cortex 5 d; Root elongation zone: root elongation zone 5 d; Root meristem zone 5 d: root meristem zone 5 d; Primary root 5 d: main root 5 d; Secondary root 7~8 d: secondary root 7~8 d; Internode 6~7: 6~7 internode; Internode 7~8: 7~8 internode; Leaf zone 1 symmetrical: leaf zone 1 (symmetrical); Leaf zone 2 stomal: Leaf area 2 (stomata); Leaf zone 3 growth: leaf zone 3 (growth); Mature leaf: mature leaf; Ear primordium 2~4 mm: ear primordium 2~4 mm; Ear primordium 6~8 mm: ear primordium 6~8 mm; Vegetative meristem 16~19 d: plant meristem 16~19 d.
图 5 玉米B3基因家族的胁迫应答分析
Control:对照;Cold:冷胁迫;Heat:热胁迫;Salt:盐胁迫;UV:紫外线胁迫;Mild water deficit:轻度干旱胁迫;Severe water deficit:重度干旱胁迫。
Figure 5. Stress response analysis of B3 gene family in maize
Cold: cold stress; Heat: heat stress; Salt: salt stress; UV: UV stress; Mild water deficit: mild drought stress: severe water deficit: severe drought stress.
表 1 玉米B3基因家族信息
Table 1 Family information associated with B3 gene in maize
基因名称
Gene
name基因ID
Gene ID染色体
Chromosome位置
Location蛋白长度
Amino acid
length/aa氨基酸分子质量
Protein molecular
weight/kDa等电点
Isoelectric
point信号肽
Signal
peptide亚细胞定位
Subcellular
localizationZmABI1 Zm00001eb143690 Chr.3 164992981-164997947 691 73.25 6.25 No Nuclear ZmABI2 Zm00001eb259870 Chr.6 5531130-5534996 317 35.05 7.09 No Chloroplast ZmABI3 Zm00001eB361390 Chr.8 158727000-158731148 392 42.64 10.14 No Chloroplast ZmARF1 Zm00001eb029770 Chr.1 162618592-162620659 446 48.73 8.66 No Cytoplasmic ZmARF10 Zm00001eb101720 Chr.2 194575898-194576928 105 11.92 11.02 No Cytoplasmic ZmARF11 Zm00001eb118970 Chr.3 1675067-1679757 684 76.59 6.29 No Nuclear ZmARF12 Zm00001eb133000 Chr.3 94801561-94808087 830 91.69 6.70 No Nuclear ZmARF13 Zm00001eb136130 Chr.3 125302568-125308760 888 97.84 6.08 No Nuclear ZmARF14 Zm00001eb142540 Chr.3 158783741-158788605 805 89.80 6.25 No Nuclear ZmARF15 Zm00001eb152940 Chr.3 201732015-201737265 720 78.34 7.17 No Nuclear ZmARF16 Zm00001eb157270 Chr.3 215856048-215864377 622 68.16 6.93 No Nuclear ZmARF17 Zm00001eb170540 Chr.4 25420464-25426332 811 89.96 7.33 No Nuclear ZmARF18 Zm00001eb182260 Chr.4 123531034-123536559 465 51.97 6.89 No Nuclear ZmARF19 Zm00001eb185290 Chr.4 148952420-148955210 748 79.75 11.10 No Nuclear ZmARF2 Zm00001eb031700 Chr.1 177503771-177511926 897 98.42 6.23 No Nuclear ZmARF20 Zm00001eb202300 Chr.4 220668443-220669476 106 12.10 11.31 No Cytoplasmic ZmARF21 Zm00001eb207450 Chr.4 245046366-245052434 847 93.91 6.16 No Nuclear ZmARF22 Zm00001eb221620 Chr.5 29000492-29005177 644 70.96 6.97 No Nuclear ZmARF23 Zm00001eb224680 Chr.5 43789840-43796267 913 100.91 6.47 No Nuclear ZmARF24 Zm00001eb225570 Chr.5 50935637-50939357 725 78.46 7.04 No Nuclear ZmARF25 Zm00001eb227100 Chr.5 59295296-59302607 1 152 127.82 6.72 No Nuclear ZmARF26 Zm00001eb232120 Chr.5 80470636-80479100 1 147 127.24 6.08 No Chloroplast ZmARF27 Zm00001eb243930 Chr.5 177184418-177190185 673 74.66 6.39 No Nuclear ZmARF28 Zm00001eb247720 Chr.5 190712368-190715567 541 57.43 6.73 No Cytoplasmic ZmARF29 Zm00001eb273540 Chr.6 98831655-98835876 700 75.51 7.56 No Nuclear ZmARF3 Zm00001eb035640 Chr.1 194732533-194747737 1 039 115.70 6.74 Yes Nuclear ZmARF30 Zm00001eb273840 Chr.6 101005067-101012686 665 73.65 6.51 No Nuclear ZmARF31 Zm00001eb292830 Chr.6 170115679-170122390 582 64.12 7.46 No Nuclear ZmARF32 Zm00001eb295830 Chr.6 175975687-175981396 659 71.99 7.77 No Nuclear ZmARF33 Zm00001eB363810 Chr.8 166328792-166334593 812 90.88 6.57 No Nuclear ZmARF34 Zm00001eB370810 Chr.8 180707713-180714633 571 62.70 8.21 No Nuclear ZmARF35 Zm00001eB373970 Chr.9 12448248-12460296 957 106.56 6.86 No Nuclear ZmARF36 Zm00001eb408800 Chr.10 13673999-13679982 813 89.70 7.25 No Nuclear ZmARF37 Zm00001eb426740 Chr.10 133561653-133564154 588 63.90 8.77 No Nuclear ZmARF38 Zm00001eb433020 Chr.10 149374022-149380403 945 103.84 6.32 No Nuclear ZmARF39 Zm00001eb433460 Chr.10 149976350-149985703 748 83.71 6.42 No Nuclear ZmARF4 Zm00001eb045640 Chr.1 236387537-236391953 681 74.54 7.92 No Nuclear ZmARF40 Zm00001eb434380 Chr.10 151402521-151404374 462 50.56 5.41 No Cytoskeleton ZmARF5 Zm00001eb066640 Chr.2 2524918-2533246 752 83.91 6.23 No Nuclear ZmARF6 Zm00001eb067270 Chr.2 3630367-3636867 930 102.17 6.19 No Nuclear ZmARF7 Zm00001eb076420 Chr.2 26933847-26940106 513 55.45 6.4 No0 Cytoplasmic ZmARF8 Zm00001eb077170 Chr.2 29481074-29481902 255 27.31 8.00 No Chloroplast ZmARF9 Zm00001eb082150 Chr.2 49388328-49393787 661 73.29 6.14 No Nuclear ZmHSI1 Zm00001eb106940 Chr.2 211825956-211838415 955 105.22 8.41 No Chloroplast ZmHSI2 Zm00001eb111570 Chr.2 223781937-223797411 793 87.23 5.84 No Nuclear ZmHSI3 Zm00001eB324300 Chr.7 166725324-166733527 957 105.24 8.42 No Chloroplast ZmHSI4 Zm00001eB330690 Chr.7 182217234-182223329 963 105.14 6.80 No Chloroplast ZmRAV1 Zm00001eb156040 Chr.3 212115620-212117266 389 40.49 9.92 No Cytoplasmic ZmRAV2 Zm00001eb294830 Chr.6 174089156-174090850 406 42.89 10.32 No Chloroplast ZmRAV3 Zm00001eB342850 Chr.8 65961433-65962833 375 40.28 10.48 No Cytoplasmic ZmRAV4 Zm00001eB360750 Chr.8 156425137-156426706 405 41.97 9.62 No Chloroplast ZmREM1 Zm00001eb001550 Chr.1 4613829-4616572 327 36.03 10.02 No Nuclear ZmREM10 Zm00001eb051370 Chr.1 260998593-261002924 809 90.56 8.92 No Nuclear ZmREM11 Zm00001eb073010 Chr.2 15684088-15686128 409 43.86 6.13 No Nuclear ZmREM12 Zm00001eb088080 Chr.2 106251779-106258244 542 60.72 10.42 No Nuclear ZmREM13 Zm00001eb124740 Chr.3 19549253-19550940 378 40.33 10.07 No Nuclear ZmREM14 Zm00001eb144270 Chr.3 170522446-170526238 693 76 549.20 6.13 No Nuclear ZmREM15 Zm00001eb161000 Chr.3 227546140-227548914 244 27.69 6.88 No Nuclear ZmREM16 Zm00001eb166640 Chr.4 6162714-6163412 232 26.32 6.75 No Cytoplasmic ZmREM17 Zm00001eb168870 Chr.4 16067079-16070427 503 55.39 10.13 No Chloroplast ZmREM18 Zm00001eb171080 Chr.4 28433030-28440841 232 26.66 8.93 No Nuclear ZmREM19 Zm00001eb187370 Chr.4 161654747-161662833 420 45.23 9.69 No Nuclear ZmREM2 Zm00001eb006360 Chr.1 17899920-17901860 239 27.08 9.50 No Nuclear ZmREM20 Zm00001eb197370 Chr.4 195899273-195901016 307 34.54 7.24 No Nuclear ZmREM21 Zm00001eb218990 Chr.5 19018569-19021152 333 37.50 10.12 No Nuclear ZmREM22 Zm00001eb219020 Chr.5 19067468-19070743 522 57.61 7.29 No Nuclear ZmREM23 Zm00001eb250760 Chr.5 204583184-204586327 422 44.97 7.93 No Nuclear ZmREM24 Zm00001eb268680 Chr.6 65750968-65751648 226 25.47 10.33 No Cytoplasmic ZmREM25 Zm00001eb290900 Chr.6 166115906-166120702 278 31.47 9.63 No Nuclear ZmREM26 Zm00001eB347830 Chr.8 98423808-98424587 259 29.34 9.80 No Nuclear ZmREM27 Zm00001eB361750 Chr.8 159865221-159870386 708 80.49 8.41 No Nuclear ZmREM28 Zm00001eB393690 Chr.9 131280885-131282501 277 29.87 10.42 No Nuclear ZmREM29 Zm00001eb401610 Chr.9 156441390-156442035 169 19.09 6.78 No Nuclear ZmREM3 Zm00001eb022070 Chr.1 84498120-84499710 273 29.65 10.89 No Nuclear ZmREM30 Zm00001eb406950 Chr.10 5668101-5669451 283 31.02 8.03 No Nuclear ZmREM31 Zm00001eb412120 Chr.10 44918969-44919649 226 25.53 9.97 No Nuclear ZmREM32 Zm00001eb413890 Chr.10 66245954-66259521 259 29.58 9.21 No Cytoplasmic ZmREM33 Zm00001eb414160 Chr.10 68351628-68352485 285 31.82 8.64 No Chloroplast ZmREM34 Zm00001eb415750 Chr.10 79426434-79427535 249 27.13 6.72 No Nuclear ZmREM35 Zm00001eb419760 Chr.10 102880836-102884729 435 48.58 10.48 No Chloroplast ZmREM36 Zm00001eb421100 Chr.10 111761405-111762472 159 17.32 8.78 No Chloroplast ZmREM37 Zm00001eb427970 Chr.10 137229874-137233357 404 43.36 7.09 No Nuclear ZmREM4 Zm00001eb030510 Chr.1 168954998-168957929 356 40.69 9.04 No Nuclear ZmREM5 Zm00001eb033180 Chr.1 184762763-184764162 291 30.60 7.64 No Chloroplast ZmREM6 Zm00001eb051330 Chr.1 260844051-260846164 324 36.87 8.66 No Nuclear ZmREM7 Zm00001eb051340 Chr.1 260876137-260878512 337 38.84 10.57 No Nuclear ZmREM8 Zm00001eb051350 Chr.1 260883320-260886825 332 38.01 10.44 No Nuclear ZmREM9 Zm00001eb051360 Chr.1 260939215-260942030 504 57.08 6.50 No Nuclear Nuclear:细胞核;Chloroplast:叶绿体;Cytoplasmic:细胞质;Cytoskeleton:细胞骨架。 -
[1] SASNAUSKAS G, MANAKOVA E, LAPĖNAS K, KAUNECKAITĖ K, SIKSNYS V. DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. Febs Journal, 2018, 285(21): 4041-4059. doi: 10.1111/febs.14649
[2] WANG W B, AO T, ZHANG Y Y, WU D, XU W, HAN B, LIU A Z. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis L.). Plant Diversity, 2021, 44(2): 201-212.
[3] BARRETO H G, SÁGIO S A, JÚNIOR A C, FEVEREIRO P, BENEDITO V A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. Plant Cell Tissue and Organ Culture, 2019, 139(2): 327-337. doi: 10.1007/s11240-019-01687-3
[4] WANG Y, DENG D, ZHANG R, WANG S, BIAN Y, YIN Z. Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Molecular Biology Reports, 2012, 39(5): 6267-6282. doi: 10.1007/s11033-012-1448-8
[5] 刘颖慧, 董志平. 植物B3转录因子的结构和功能. 分子植物育种, 2017, 15(5): 1868-1873. LIU Y H, DONG Z P. Structure and function of plant B3 transcription factor. Molecular Plant Breeding, 2017, 15(5): 1868-1873.
[6] ROMANEL E A, SCHRAGO C G, COUÑAGO R M, RUSSO C A, ALVES-FERREIRA M. Evolution of the B3 DNA binding superfamily, new insights into rem family gene diversification. PLoS One, 2009, 4(6): 1-18.
[7] 吴佳军, 俞率成, 刘志刚, 傅鹰, 周明兵. 毛竹B3家族全基因组鉴定及表达模式分析. 农业生物技术学报, 2019, 27(1): 43-54. WU J J, YU S C, LIU Z G, FU Y, ZHOU M B. Genome wide identification and expression pattern analysis of pubescens B3 family in Phyllostachys. Journal of Agricultural Biotechnology, 2019, 27(1): 43-54.
[8] VERMA S, BHATIA S. A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum L.). 3 Biotech, 2019, 9(9): 1-17.
[9] CHANDLER J W. Auxin response factors. Plant Cell and Environment, 2016, 39(5): 1014-1028. doi: 10.1111/pce.12662
[10] LI S B, XIE Z Z, HU C G, ZHANG J Z. A review of auxin response factors (ARFs) in plants. Frontiers in Plant Science, 2016, 7(1): 47-48.
[11] LIU Z, MIAO L, HUO R, SONG X, JOHNSON C, KONG L, SUNDARESAN V, YU X. ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant and Cell Physiology, 2018, 59(1): 179-189.
[12] WILMOTH J C, WANG S, TIWARI S B, JOSHI A D, HAGEN G, GUILFOYLE T J, ALONSO J M, ECKER J R, REED J W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal, 2005, 43(1): 118-130. doi: 10.1111/j.1365-313X.2005.02432.x
[13] GUO F, ZHANG P, WU Y, LIAN G, YANG Z, LIU W, BUERTE B, ZHOU C, ZHANG W, LI D, HAN N, TONG Z, ZHU M, XU L, CHEN M, BIAN H. Rice LEAFY COTYLEDON1 hinders embryo greening during the seed development. Frontiers in Plant Science, 2022, 13(1): 1-18.
[14] FREITAS N C, BARRETO H G, TORRES L F, FREIRE L L, RODRIGUES L, DINIZ L, BEIJO L A. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica, competence acquisition and developmental marker genes. Plant Cell Tissue and Organ Culture, 2019, 137(3): 599-611. doi: 10.1007/s11240-019-01594-7
[15] ZINSMEISTER J, LEPRINCE O, BUITINK J. Molecular and environmental factors regulating seed longevity. Biochemical Journal, 2020, 477(2): 305-323. doi: 10.1042/BCJ20190165
[16] KHANDELWAL A, CHO S H, MARELLA H, SAKATA Y, PERROUD P F, PAN A, QUATRANO R S. Role of ABA and ABI3 in desiccation tolerance. Science, 2010, 327: 546-547. doi: 10.1126/science.1183672
[17] 张一弓, 张怡, 阿依白合热木·木台力甫, 张道远. 异源过表达齿肋赤藓ScABI3基因改变拟南芥气孔表型并提高抗旱性. 植物学报, 2021, 56(4): 414-421. doi: 10.11983/CBB20212 ZHANG Y G, ZHANG Y, Ayibaiheremu·Mutailifu, ZHANG D Y. Heterologous overexpression of ScABI3 gene in Erythritus denticola alters the stomatal phenotype of Arabidopsis thaliana and improves drought resistance. Acta Botanica Sinica, 2021, 56(4): 414-421. doi: 10.11983/CBB20212
[18] KIM H U, JUNG S J, LEE K R, KIM E H, LEE S M, ROH K H, KIM J B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio, 2014, 4(1): 25-32. doi: 10.1016/j.fob.2013.11.003
[19] VEERAPPAN V, WANG J, KANG M, LEE J, TANG Y, JHA A K, SHI H, PALANIVELU R, ALLEN R D. A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to depression of seed-specific gene expression. Planta, 2012, 236(1): 1-17. doi: 10.1007/s00425-012-1630-1
[20] CHEN N, VEERAPPAN V, ABDELMAGEED H, KANG M, ALLEN RD. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell, 2018, 30(3): 600-619. doi: 10.1105/tpc.17.00655
[21] LI X J, LI M, ZHOU Y, HU S, HU R, CHEN Y, LI X B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One, 2015, 10(2): 1-18.
[22] PENG Z, WANG M, ZHANG L, JIANG Y, ZHAO C, SHAHID M Q, BAI Y, HAO J, PENG J, GAO Y, SU W, YANG X. EjRAV1/2 Delay Flowering Through Transcriptional Repression of EjFTs and EjSOC1s in Loquat. Frontiers in Plant Science, 2021, 12(1): 1-14.
[23] KING G J, CHANSON A H, MCCALLUM E J, OHME-TAKAGI M, BYRIEL K, HILL J M, MARTIN J L, MYLNE J S. The Arabidopsis B3 domain protein VERNALIZATION1 (VRN1) is involved in processes essential for development, with structural and mutational studies revealing its DNA-binding surface. Journal of Biological Chemistry, 2013, 288(5): 3198-3207. doi: 10.1074/jbc.M112.438572
[24] CASELLI F, BERETTA V M, MANTEGAZZA O, PETRELLA R, LEO G, GUAZZOTTI A, HERRERA-UBALDO H, DE FOLTER S, MENDES M A, KATER M M, GREGIS V. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Frontiers in Plant Science, 2019, 10(2): 1351-1368.
[25] WANG W, ZHANG X, DENG F, YUAN R, SHEN F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genomics, 2017, 18(1): 376-401. doi: 10.1186/s12864-017-3768-5
[26] WARMAN C, PANDA K, VEJLUPKOVA Z, HOKIN S, UNGER-WALLACE E, COLE R A, CHETTOOR A M, JIANG D, VOLLBRECHT E, EVANS M M S, SLOTKIN R K, FOWLER J E. High expression in maize pollen correlates with genetic contributions to pollen fitness as well as with coordinated transcription from neighboring transposable elements. PLoS Genetics, 2020, 16(4): 1-32.
[27] WALLEY J W, SARTOR R C, SHEN Z, SCHMITZ R J, WU K J, URICH M A, NERY J R, SMITH L G, SCHNABLE J C, ECKER J R, BRIGGS S P. Integration of omic networks in a developmental atlas of maize. Science, 2016, 353: 814-818. doi: 10.1126/science.aag1125
[28] MAKAREVITCH I, WATERS A J, WEST P T, STITZER M, HIRSCH C N, ROSS-IBARRA J, SPRINGER N M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genetics, 2015, 11(1): 1-12.
[29] OPITZ N, PASCHOLD A, MARCON C, MALIK W A, LANZ C, PIEPHO H P, HOCHHOLDINGER F. Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genomics, 2014, 15(1): 741-754. doi: 10.1186/1471-2164-15-741
[30] 石荣康, 张冬梅, 孙正文, 刘正文, 解美霞, 张艳, 马峙英, 王省芬. 陆地棉REM基因家族全基因组鉴定及表达分析. 棉花学报, 2021, 33(2): 95-111. doi: 10.11963/1002-7807.srkwxf.20210113 SHI R K, ZHANG D M, SUN Z W, LIU Z W, XIE M X, ZHANG Y, MA S Y, WANG S F. Complete genome identification and expression analysis of REM gene family in Gossypium hirsutum. Cotton Science, 2021, 33(2): 95-111. doi: 10.11963/1002-7807.srkwxf.20210113
[31] 孙亭亭, 王大伟, 龚达平, 陈乐, 陈雅琼, 孙玉合. 番茄B3超家族成员鉴定及生物信息学分析. 植物遗传资源学报, 2015, 16(4): 806-814. SUN T T, WANG D W, GONG D P, CHEN L, CHEN Y Q, SUN Y H. Identification and bioinformatics analysis of tomato B3 superfamily members. Journal of Plant Genetic Resources, 2015, 16(4): 806-814.
[32] 关思静, 高静, 徐蓉蓉, 葛甜甜, 王楠, 颜永刚, 张岗, 陈莹, 刘阿萍, 程萌格. 甘草生长素反应因子(ARF)基因家族的鉴定及表达分析. 中国农学通报, 2021, 37(29): 20-27. doi: 10.11924/j.issn.1000-6850.casb2020-0304 GUAN S J, GAO J, XU R R, GE T T, WANG N, YAN Y G, ZHANG G, CHEN Y, LIU A P, CHENG M G. Identification and expression analysis of auxin response factor (ARF) gene family in Glycyrrhiza uralensis. Chinese Agricultural Science Bulletin, 2021, 37(29): 20-27. doi: 10.11924/j.issn.1000-6850.casb2020-0304
[33] 李庆飞, 王梦梦, 郭卫丽, 周俊国, 李新峥. 南瓜转录因子基因CmRAV的克隆与表达分析. 植物生理学报, 2021, 57(12): 2337-2345. doi: 10.13592/j.cnki.ppj.2021.0144 LI Q F, WANG M M, GUO W L, ZHOU J G, LI X Z. Cloning and expression analysis of pumpkin transcription factor gene CmRAV. Chinese Journal of Plant Physiology, 2021, 57(12): 2337-2345. doi: 10.13592/j.cnki.ppj.2021.0144
[34] 瓮巧云, 赵彦敏, 张贺, 宋晋辉, 马海莲, 袁进成, 刘颖慧. 玉米ZmREM基因的克隆与逆境胁迫后表达分析. 浙江农业学报, 2016, 28(11): 1822-1827. doi: 10.3969/j.issn.1004-1524.2016.11.03 WENG Q Y, ZHAO Y M, ZHANG H, SONG J H, MA H L, YUAN J C, LIU Y H. Cloning and expression analysis of ZmREM gene in maize. Acta Agriculturae Zhejiangensis, 2016, 28(11): 1822-1827. doi: 10.3969/j.issn.1004-1524.2016.11.03
[35] ZHAO S P, XU Z S, ZHENG W J, ZHAO W, WANG Y X, YU T F, CHEN M, ZHOU Y B, MIN D H, MA Y Z, CHAI S C, ZHANG X H. Genome-wide analysis of the RAV family in soybean and functional identification of GMRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Frontiers in Plant Science, 2017, 6(8): 905-916.
[36] 闵浩巍. 玉米抗逆相关基因ZmRAV1和ZmTCX8.1的功能研究. 北京: 中国农业大学博士学位论文, 2013. MIN H W. Study on the function of maize stress resistance related genes ZmRAV1 and ZmTCX8.1. PhD Thesis. Beijing: China Agricultural University, 2013.
[37] WENG Q Y, S ONG J H, MA H L, YUAN J C, ZHAO Y M, ZHAO Y, LIU Y H. Cloning and expression analysis of ZmABI3 gene in Zea mays. Turkish Journal of Biochemistry, 2017, 42(3): 279-285.
-
期刊类型引用(0)
其他类型引用(1)



 下载:
下载: