大颖草和短芒披碱草的染色体FISH分析
大颖草(Kengyilia grandiglumis)和短芒披碱草(Elymus breviaristatus)是分布在青藏高原沙生环境中的两种多年生禾草。本研究采用顺序荧光原位杂交(FISH)和基因组原位杂交(GISH)技术,应用转座子探针S5和串联重复序列探针AAG对两禾草根尖有丝分裂中期的染色体进行了分析。结果显示,GISH信号可成功区分P、H、St和Y 4个亚基因组,在此基础上获得了S5和AAG探针在两种植物21对染色体上的分布信息。大颖草中S5信号多分布在端粒及近端粒区,P亚基因组中分布位点最多;AAG信号多分布在近着丝粒区和近端粒区, Y亚基因组中分布的位点最多。短芒披碱草中S5信号分布在端粒、近端粒区、近着丝粒区和臂中部区,H亚基因组上信号比St和Y亚基因组丰富;AAG信号分布在近着丝粒区、臂中部区和近端粒区,信号分布位点在St亚基因组中最少。本研究获得了两种异源六倍体禾草亚基因组水平的染色体信号特征,为大颖草和短芒披碱草种质资源的评价及利用奠定了基础。
English
-
青藏高原由于其独特的自然环境,孕育了许多代表性植物类群。其中就有两种生长在荒漠草原中的禾草大颖草(Kengyilia grandiglumis)和短芒披碱草(Elymus breviaristatus)。大颖草隶属于禾本科小麦族以礼草属,为多年生丛生草本植物,主要分布于青海省海拔2 300~4 100 m的沙丘、山坡、草地等生境中[1-2]。短芒披碱草隶属于禾本科小麦族披碱草属,为多年生疏丛草本植物,主要分布在四川和青海等省山坡生境中[1-2]。大颖草和短芒披碱草具有适应性强和产量高等优点,是高寒地区栽培草地建植和生态修复较为理想的牧草草种[3-4]。
围绕大颖草和短芒披碱草,国内外学者开展了以下不同层面的研究。核型分析及荧光原位杂交报道大颖草为异源六倍体(2n = 6x = 42),其基因组组成为StPY [5-6]。利用叶绿体基因和核基因探讨了大颖草母本供体来源及其与其他以礼草属物种的系统关系[7-8]。此外,应用随机扩增微卫星多态性(random amplified microsatellite polymorphism, RAMP)、EST-SSR和荧光原位杂交(fluorescence in situ hybridization,FISH)标记分别评估了大颖草的遗传多样性[9-11],研究结果表明在以礼草属物种中,大颖草遗传多样性偏低[11]。短芒披碱草的核型分析报道其为异源六倍体(2n = 6x = 42),基因组组成为StHY [12-13]。分别利用形态学标记、SRAP和EST-SSR分子标记对短芒披碱草开展了遗传多样性和群体遗传结构的研究,研究结果表明短芒披碱草表型多样性丰富,自然居群间遗传分化较大,但其居群间及居群内遗传多样性远低于同属其他物种[14-17]。
FISH的基本原理是根据核酸分子碱基互补配对的原则,将变性后带有荧光标记的探针与变性后的靶核酸序列在退火温度下复性,通过荧光显微镜等仪器观察杂交信号[18]。由于荧光原位杂交能将基因组中特定DNA序列与染色体结构及组织定位直接联系起来,提供分子生物学层面不能揭示的生物遗传信息,因此被广泛应用于植物研究中的染色体识别、基因定位、图谱构建和系统进化分析等领域[19-23]。
大颖草和短芒披碱草均为狭域分布在青藏高原的特有小麦族物种,由于其种群较小和人类活动加剧环境恶化等因素影响,这两物种的生存受到了威胁。如短芒披碱草曾被列入第一批国家重点野生保护植物名录中,属于国家二级保护植物。本研究选取大颖草和短芒披碱草作为研究对象,采用荧光原位杂交技术对两种异源六倍体物种进行染色体识别和核型分析,探究其在染色体水平的遗传多样性,以期为大颖草和短芒披碱草种质资源的合理保护、评价及利用提供理论依据。
1. 材料与方法
1.1 试验材料
大颖草和短芒披碱草均采自位于青海省海晏县的国家小麦野生近缘植物原生境保护区(36°50′ N,100°50′ E),该区位于青海湖东北部环湖沙化地带,平均海拔3 280 m [24]。间隔10 m分别随机选取5株大颖草和短芒披碱草作为试验对象。糙缘拟鹅观草(Pseudoroegneria stipifolia,PI 313960)、布顿大麦(Hordeum bogdanii,PI 440413)和沙芦草(Agropyron mongolicum,PI 499392)来源于美国农业部的国家植物种质资源库(NPGS),均为二倍体物种。
1.2 试验方法
1.2.1 染色体制备
将采集的种子于实验室条件萌发,待根长至0.5~1 cm时取其根尖,在N2O中处理2 h [25]。处理结束后,向每管中加入卡诺固定液(乙醇 ꞉ 乙酸= 3 ꞉ 1)在4 ℃固定5 min以上。固定后的根尖用45%的醋酸压片。在显微镜下观察,将具有分裂相多、染色体分散、染色体形态清晰的片子置于−80 ℃保存30 min以上。用刀片除去盖玻片后,将载玻片风干以作进一步处理。
1.2.2 FISH和GISH探针的制备
用转座子(transposable elements,TEs)和串联重复序列作为FISH标记来区分大颖草和短芒披碱草的染色体。S5是从St基因组中开发的DNA转座子探针[26],AAG是被广泛应用于小麦族的微卫星探针[27]。S5和(AAG)10寡核苷酸探针在上海生工生物公司合成分别具有5′修饰荧光基团TAMRA和FAM。GISH探针标记参考Dou等[28]报道的方法,分别将糙缘拟鹅观草、布顿大麦和沙芦草的基因组DNA片段化后用随机引物标记法标记。
1.2.3 荧光原位杂交
所有材料均采用顺序FISH和GISH进行两轮荧光原位杂交。荧光原位杂交流程按照Liu 等[29]方法。杂交后用DAPI (4',6-二脒基-2-苯基吲哚)复染,最后在Olympus BX53荧光显微镜下观察图像,并使用Adobe Photoshop 6.0进行对比度和亮度调节。每株材料至少观察和统计3个细胞的染色体FISH结果。
2. 结果与分析
2.1 大颖草根尖细胞染色体FISH分析
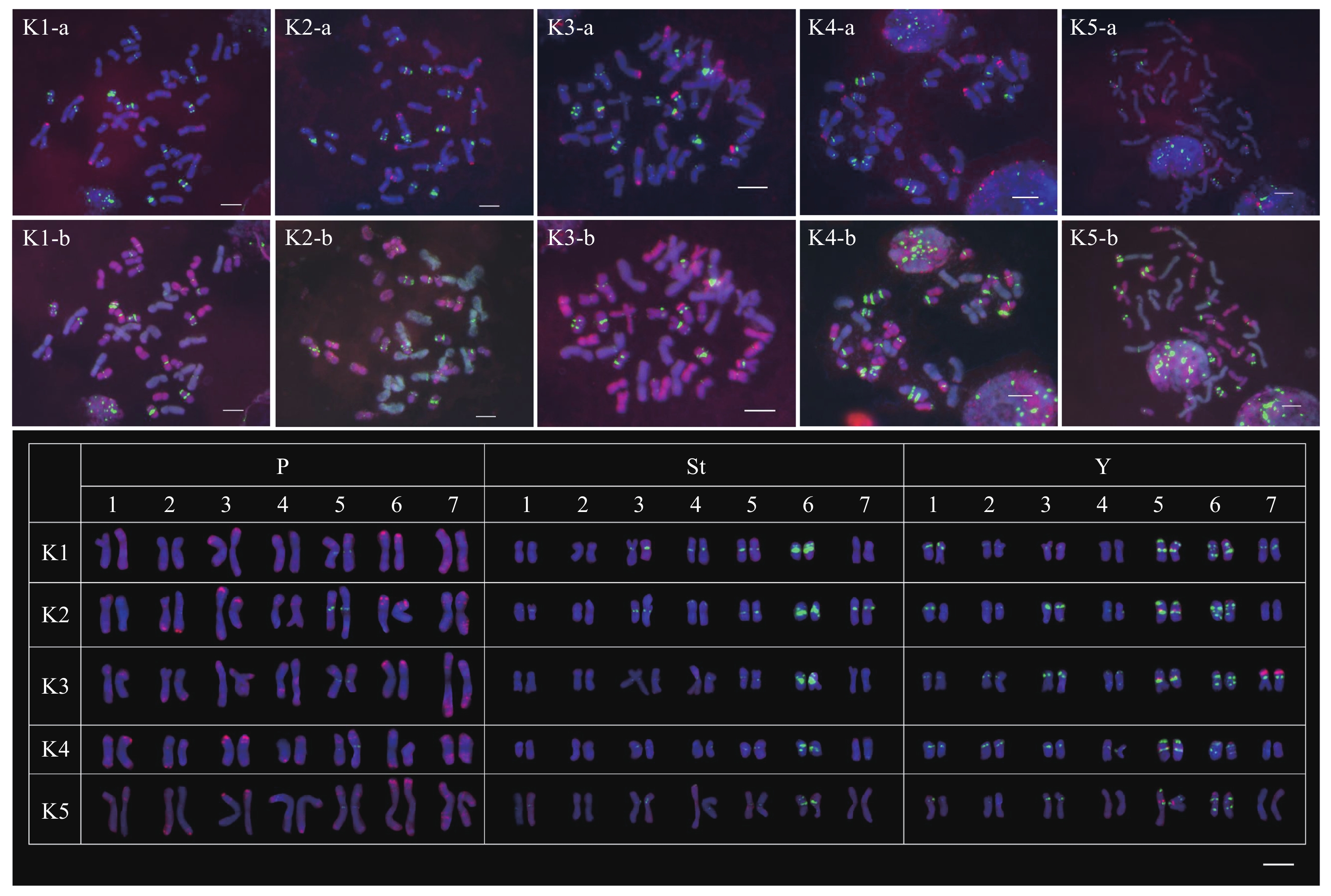
本研究采集的5株大颖草(K1~K5)染色体数目均为2n = 6x = 42,所有染色体根据第2轮GISH信号可划分为P、St和Y 3个亚基因组,不同亚基因组间未发现易位现象。每个亚基因组中的染色体根据臂比、相对长度和探针信号按1~7排列(图1)。
![]() 图 1 顺序FISH和GISH探针在大颖草体细胞染色体上的荧光原位杂交结果K1-a、K2-a、K3-a、K4-a和K5-a为第1轮S5 (红色)和(AAG)10 (绿色)探针的杂交信号。K1-b、K2-b、K3-b、K4-b和K5-b为第2轮糙缘拟鹅观草(红色)和沙芦草(绿色)基因组DNA的杂交信号。Figure 1. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Kengyilia grandiglumis.K1-a, K2-a, K3-a, K4-a, and K5-a: initially FISH probed with S5 (red) and (AAG)10 (green). K1-b, K2-b, K3-b, K4-b, and K5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Agropyron mongolicum (green).
图 1 顺序FISH和GISH探针在大颖草体细胞染色体上的荧光原位杂交结果K1-a、K2-a、K3-a、K4-a和K5-a为第1轮S5 (红色)和(AAG)10 (绿色)探针的杂交信号。K1-b、K2-b、K3-b、K4-b和K5-b为第2轮糙缘拟鹅观草(红色)和沙芦草(绿色)基因组DNA的杂交信号。Figure 1. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Kengyilia grandiglumis.K1-a, K2-a, K3-a, K4-a, and K5-a: initially FISH probed with S5 (red) and (AAG)10 (green). K1-b, K2-b, K3-b, K4-b, and K5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Agropyron mongolicum (green).在大颖草中,S5信号分布在染色体端粒、近端粒区、近着丝粒区和臂中部区。在P亚基因组中,1~7P染色体上均观察到了S5信号。除2P染色体短臂端粒区无信号外,S5信号在其余染色体的端粒或近端粒区呈现出数量和强弱的差异。7P染色体是P亚基因组中S5信号分布位点最多的染色体。与P亚基因组相比,S5信号在St和Y亚基因组中较少且微弱。St亚基因组中,仅在部分2St、4St和7St染色体的近端粒区,5St染色体的近着丝粒区产生了少量的杂交信号(图1)。Y亚基因组中,5株材料的5Y染色体短臂的近着丝粒区和6Y染色体短臂的端粒区观察到S5信号。值得注意的是7Y染色体,仅在1份材料的短臂端粒区观察到强S5信号(图1 K3)。
(AAG)10信号分布在Y、St和P亚基因组染色体的近着丝粒区、臂中部区和端粒区。与S5相比,(AAG)10信号在端粒及近端粒区的分布较少。整体来看,(AAG)10信号在Y亚基因组中分布的位点最多,而在P亚基因组中最少。在P亚基因组中仅观察到5P染色体近着丝粒区的(AAG)10信号。在St亚基因组中,除了2St染色体,其余染色体上均有AAG信号的杂交位点,且大多分布在近着丝粒区(图1)。K1、K2和K3材料的6St染色体长臂近着丝粒区的(AAG)10信号尤为强烈。Y亚基因组的1~7号染色体上均有不同强度及数量的(AAG)10信号分布,但不同染色体间信号变化差异大。5Y和6Y染色体上(AAG)10信号分布位置固定,仅部分材料中强度有变化。而7St染色体(AAG)10信号变化大,K1和K3材料中观察到位于短臂近着丝粒区的信号,但在其他3份材料中未观察到信号(图1)。
2.2 短芒披碱草根尖细胞染色体FISH分析
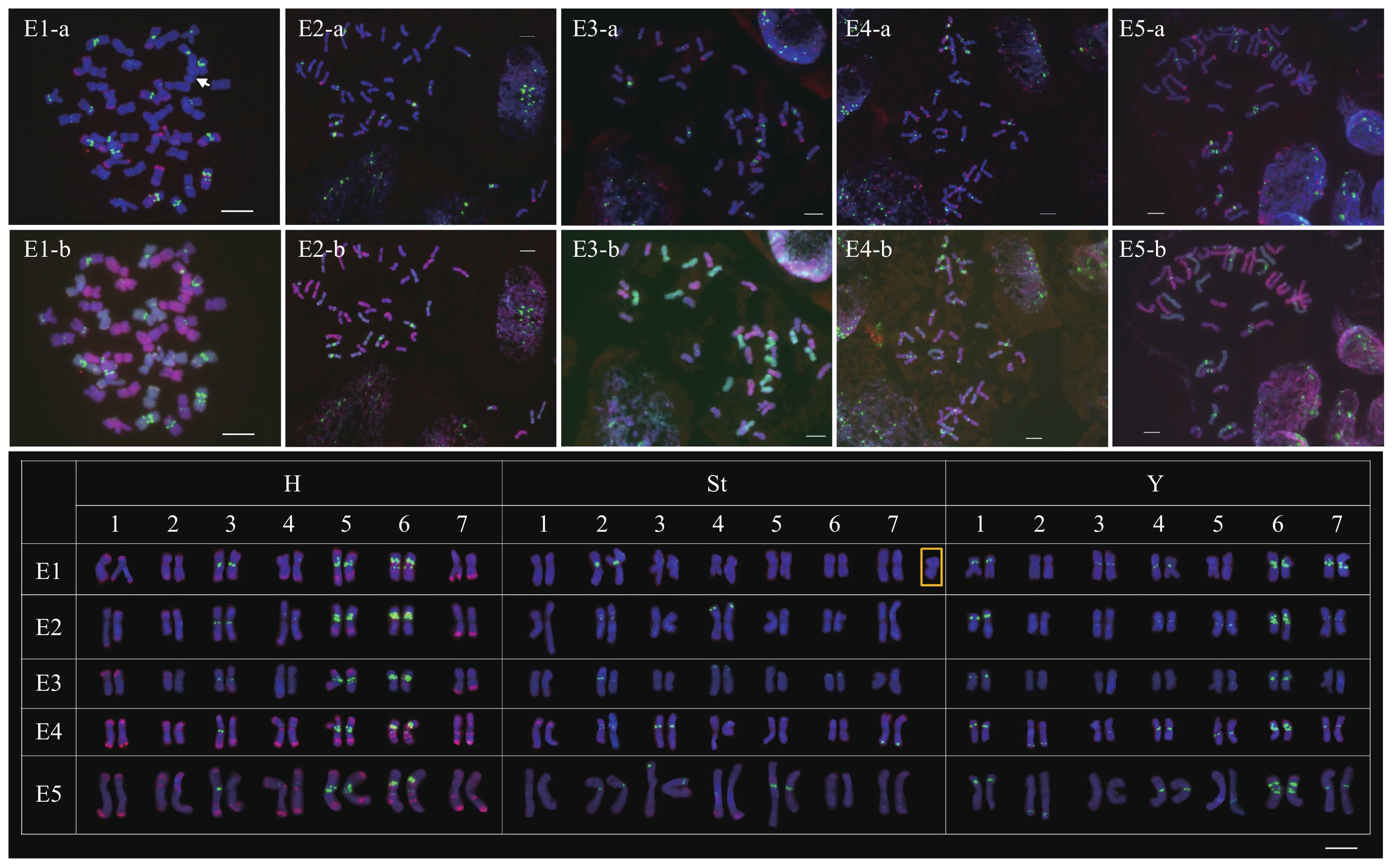
在采集的5株短芒披碱草(E1~E5)中,1株染色体数目为43条(E1),其余为42条。所有染色体根据第2轮GISH信号可划分为H、St和Y 3个亚基因组,不同亚基因组间未发现易位现象。每个亚基因组中的染色体根据臂比、相对长度和探针信号按1~7排列(图2)。根据GISH信号判定E1材料中多出的一条染色体属于St亚基因组(图2 E1-b)。
![]() 图 2 顺序FISH和GISH探针在短芒披碱草体细胞染色体上的荧光原位杂交结果E1-a、E2-a、E3-a、E4-a和E5-a为第1轮S5 (红色)和AAG (绿色)探针的杂交信号。E1-b、E2-b、E3-b、E4-b和E5-b为第2轮糙缘拟鹅观草(红色)和布顿大麦(绿色)基因组DNA的杂交信号。箭头和方框指示的是非整倍中的第43条染色体。Figure 2. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Elymus breviaristatus.E1-a, E2-a, E3-a, E4-a, and E5-a: initially FISH probed with S5 (red) and (AAG) 10 (green). E1-b, E2-b, E3-b, E4-b, and E5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Hordeum bogdanii (green). The arrow and box indicate chromosome 43 in aneuploid.
图 2 顺序FISH和GISH探针在短芒披碱草体细胞染色体上的荧光原位杂交结果E1-a、E2-a、E3-a、E4-a和E5-a为第1轮S5 (红色)和AAG (绿色)探针的杂交信号。E1-b、E2-b、E3-b、E4-b和E5-b为第2轮糙缘拟鹅观草(红色)和布顿大麦(绿色)基因组DNA的杂交信号。箭头和方框指示的是非整倍中的第43条染色体。Figure 2. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Elymus breviaristatus.E1-a, E2-a, E3-a, E4-a, and E5-a: initially FISH probed with S5 (red) and (AAG) 10 (green). E1-b, E2-b, E3-b, E4-b, and E5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Hordeum bogdanii (green). The arrow and box indicate chromosome 43 in aneuploid.S5信号分布在短芒披碱草染色体的端粒、近端粒区、近着丝粒区和臂中部区。H亚基因组S5信号比St和Y亚基因组丰富(图2)。在H亚基因组中,1~7H染色体上均有S5信号的分布,且短臂与长臂的近端粒区均能观察到不同强度的S5信号。在这5份材料中,S5信号分布位置较为保守,仅仅呈现出信号强弱的变化。与H亚基因组相比,仅在St和Y亚基因组中的个别染色体上观察到了S5信号。在St亚基因组中, E3材料的1St、3St、4St和7St染色体的近端粒区和臂中部区产生了少量的杂交信号(图2 E3);E4材料的2St、3St和4St染色体的近端粒区产生了少量的杂交信号(图2 E4)。Y亚基因组中,仅在部分6Y染色体短臂的端粒区观察到S5信号。
在短芒披碱草中,(AAG)10信号分布在H、St和Y亚基因组染色体的近着丝粒区、臂中部区和近端粒区。整体来看,(AAG)10信号在Y和H亚基因组中分布的位点较多,而在St亚基因组中最少。Y亚基因组的1~7号染色体上均有不同强度及数量的(AAG)10信号分布,1Y、3Y、4Y和6Y染色体上(AAG)10信号分布位置固定,其他染色体上有信号数量和分布位置的变化。2Y染色体近端粒区的(AAG)10信号仅在E4和E5材料中发现,其他3份材料中未观察到(AAG)10信号。与其他材料7Y染色体(AAG)10信号仅分布在短臂近着丝粒区相比,E1材料中的(AAG)10信号在7Y染色体长臂和短臂近着丝粒区均有分布。在H亚基因组中,5株材料的3H、4H、5H和6H染色体近着丝粒区观察到强度不一的(AAG)10信号。在St亚基因组中,(AAG)10信号在不同材料中变化较大。如6St染色体,仅在E3材料中观察到位于短臂端粒区的信号;7St染色体,E1、E4和E-5材料中(AAG)10信号位于长臂的近端粒区,而在E2和E3材料中分别位于短臂的臂中部区和近着丝粒区(图2)。
值得注意的是,在短芒披碱草E4和E5材料的3H染色体上观察到了(AAG)10信号的杂合,即仅在1条染色体上有位于长臂近着丝粒区的(AAG)10信号(图2 E4,E5)。
3. 讨论与结论
本研究应用TE探针S5、串联重复序列探针(AAG)10及GISH探针成功区分识别了大颖草和短芒披碱草两种异源六倍体物种的3个亚基因组的21对染色体。TEs是指弥散分布在基因组的一些重复序列,其重复单元并不相连,而是与其他序列参杂在一起。串联重复序列由含有一定碱基的重复单元首尾串联,以多次重复的方式形成阵列成簇排布于染色体端部、着丝粒周围以及染色体异染色质区域处[30]。由于TEs和串联重复序列各自特性的不同,其FISH信号也具有不同的特征。本研究中,S5探针呈现出点状至弥散分布,(AAG)10探针为点状分布。分布位置在不同基因组间也具有一定的倾向性。在P、H、St和Y亚基因组中,S5信号集中分布在端粒及近端粒区,少数分布在近着丝粒区和臂中部区;而(AAG)10信号在上述基因组中分布位点最多的是近着丝粒区,其次为近端粒区,臂中部区最少。本研究中,与H和P基因组的FISH信号相比较,St和Y基因组信号较少,在今后的研究中,应考虑开发St和Y基因组特异的FISH探针,以期获得St和Y基因组的更多信息。
大颖草和短芒披碱草中都具有St和Y亚基因组,但荧光原位杂交结果未显示出两者的S5和(AAG)10信号具有相似的规律。Yen等[31]认为含St基因组的六倍体物种经历了两次杂交事件。第1次杂交事件发生在二倍体物种间,形成了StStYY、StStPP和StStHH四倍体物种;以上四倍体物种再分别与二倍体含H、P和W等基因组的物种二次杂交,形成六倍体物种。依据大颖草和短芒披碱草St和Y亚基因组相异的FISH信号,本研究推测在大颖草和短芒披碱草的异源多倍体形成中,不同含St和Y基因组的祖先种参与了此过程。
本研究在1株短芒披碱草中发现了43条染色体即非整倍体的存在。与大多数动物物种相比,多倍体植物对非整倍性具有相当的耐受性[32]。例如,在16个新合成六倍体小麦品系中,发现非整倍性从20%到100%不等[33]。但普遍认为尽管非整倍体增加了群体的遗传多样性,但对群体进化的贡献很小或没有[34]。本研究中大颖草未发现非整倍体现象,可能是以礼草属物种特性,如在糙毛以礼草(Kengyilla hirsute)核型研究中也未发现非整倍体[35];也可能与试验材料个数较少有关,需在今后的研究中加大采样量来进一步确定。
综上所述,本研究应用TE探针、串联重复序列探针及GISH探针,分别获得了大颖草和短芒披碱草3个亚基因组及21对染色体的FISH信号特征,为多倍体禾草种质资源多样性研究及多倍体形成演化提供参考。
参考文献
[1] 中国科学院中国植物志委员会. 中国植物志. 第九卷. 北京: 科学出版社, 1987: 6-116. Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora Reipublicae Popularis Sinicae. vol. 9. Beijing: Science Press, 1987: 6-116.
[2] 青海植物志编辑委员会, 青海植物志. 第四卷. 西宁: 青海人民出版社, 1996: 87-98. Editorial Committee of Flora of Qinghai. Flora of Qinghai. vol 4. Xining: Qinghai People’s House, 1996: 87-98.
[3] 唐俊伟, 乔安海, 马力, 贾顺斌, 王晓彤. 播种技术对大颖草产量及农艺性状的影响. 草地学报, 2019, 27(5): 1425-1430. TANG J W, QIAO A H, MA L, JIA S B, WANG X T. The effect of the rowing technologies on yield and agronomic characters of Roegeria grandigumis. Acta Agrestia Sinica, 2019, 27(5): 1425-1430.
[4] 施建军, 马玉寿, 董全民, 王彦龙, 王柳英. “黑土型”退化草地优良牧草筛选试验. 草地学报, 2007, 15(6): 543-549. SHI J J, MA Y S, DONG Q M, WANG Y L, WANG L Y. The selection experiment of fine forages in ‘Black Siol Type’ degraded grasslands. Acta Agrestia Sinica, 2007, 15(6): 543-549.
[5] 张利, 周永红, 郑有良, 张颖. 仲彬草属八个物种的核型. 植物分类与资源学报, 2005, 27(1): 81-86. ZHANG L, ZHOU Y H, ZHENG Y L, ZHANG Y. Karyotypes of 8 species in Kengyilia. Acta Botanica Yunnanica, 2005, 27(1): 81-86.
[6] DOU Q W, WANG R R-C, LEI Y T, YU F, LI Y, WANG H Q, CHEN Z G. Genome analysis of seven species of Kengyilia (Triticeae: Poaceae) with FISH and GISH. Genome, 2013, 56(11): 641-649. doi: 10.1139/gen-2013-0113
[7] ZENG J, ZHANG L, FAN X, ZHANG H Q, YANG R W, ZHOU Y H. Phylogenetic analysis of Kengyilia species based on nuclear ribosomal DNA internal transcribed spacer sequences. Biologia Plantarum, 2008, 52(2): 231-236. doi: 10.1007/s10535-008-0051-2
[8] LUO X M, TINKER N A, FAN X, ZHANG H Q, SHA L N, KANG H Y, DING C B, LIU J, ZHANG L, YANG R W, ZHOU Y H. Phylogeny and maternal donor of Kengyilia species (Poaceae: Triticeae) based on three cpDNA (matK, rbcL and trnH-psbA) sequences. Biochemical Systematics and Ecology, 2012, 44: 61-69.
[9] ZHANG L, ZHENG Y L, WEI Y M, LIU S G, ZHOU Y H. The genetic diversity and similarities among Kengyilia species based on random amplified microsatellite polymorphism (RAMP). Genetic Resources and Crop Evolution, 2005, 2(8): 1011-1017.
[10] WANG Q X, XIANG J S, GAO A N, YANG X M, LIU W H, LI X Q, LIU H L. Analysis of chromosomal structural polymorphisms in the St, P, and Y genomes of Triticeae (Poaceae). Genome, 2010, 3(3): 241-249.
[11] 任建东, 李凤珍, 徐媛君, 王晓醒, 马晓岗. 基于EST-SSR分子标记的青海高原以礼草属主要物种的遗传多样性分析. 植物遗传资源学报, 2016, 17(4): 663-670. REN J D, LI F Z, XU Y J, WANG X X, MA X G. Genetic diversity of the major varieties of Kengyilia in Qinghai Plateau based on EST-SSR markers. Journal of Plant Genetic Resources, 2016, 17(4): 663-670.
[12] 刘玉红. 我国11种披碱草的核型研究. 植物科学学报, 1985, 3(4): 325-330. LIU Y H. Studies on the karyotypes of 11 species of Elymus from China. Journal of Wuhan Botanical Research, 1985, 3(4): 325-330.
[13] YANG C R, BAUM B-R, CHEN W H, ZHANG H Q, LIU X Y, FAN X, SHA L N, KANG H Y, WANG Y, ZHOU Y H. Genomic constitution and taxonomy of the Chinese hexaploids Elymus cylindricus and E. breviaristatus (Poaceae: Triticeae). Botanical Journal of the Linnean Society, 2016, 182(3): 650-657. doi: 10.1111/boj.12469
[14] 顾晓燕, 郭志慧, 张新全, 周凯, 周朝杰, 符开欣, 刘新, 马啸. 短芒披碱草异位保护群体的表型多样性研究. 草业学报, 2015, 24(5): 141-152. doi: 10.11686/cyxb20150517 GU X Y, GUO Z H, ZHANG X Q, ZHOU K, ZHOU C J, FU K X, LIU X, MA X. Phenotypic variations in seven ex-situ conservation populations of Elymus breviaristatus. Acta Prataculturae Sinica, 2015, 24(5): 141-152. doi: 10.11686/cyxb20150517
[15] GU X Y, GUO Z H, MA X, BAI S Q, ZHANG X Q, ZHANG C B, CHEN S Y, PENG Y, YAN Y H, HUANG L K, ZHOU K, ZHOU C J, FU K X. Population genetic variability and structure of Elymus breviaristatus (Poaceae: Triticeae) endemic to Qinghai-Tibetan Plateau inferred from SSR markers. Biochemical Systematics and Ecology, 2015, 58: 247-256.
[16] LI J, ZHANG C B, CHEN S Y, JIANG K K, GUAN H, LIU W H. Characterization and application of EST-SSR markers developed from transcriptome sequences in Elymus breviaristatus (Poaceae: Triticeae). Genes, 2023, 14(2): 302. doi: 10.3390/genes14020302
[17] 顾晓燕, 郭智慧, 张新全, 刘新, 周朝杰, 马啸. 短芒披碱草SRAP-PCR体系的建立和优化. 中国农学通报, 2014, 30(30): 259-264. doi: 10.11924/j.issn.1000-6850.2014-1190 GU X Y, GUO Z H, ZHANG X Q, LIU X, ZHOU C J, MA X. Establishment and optimization of SRAP-PCR reaction system for Elymus breviaristatus. Chines Agricultural Science Bulletin, 2014, 30(30): 259-264. doi: 10.11924/j.issn.1000-6850.2014-1190
[18] BLANCATO J K. Fluorescence in Situ Hybridization. In The Principles of Clinical Cytogenetics. New Jersey: Humana Press, 1999: 443-471.
[19] KATO A, VEGA J M, HAN F P, LAMB J C, BIRCHLER J A. Advances in plant chromosome identification and cytogenetic techniques. Current Opinion in Plant Biology, 2005, 8(2): 148-154. doi: 10.1016/j.pbi.2005.01.014
[20] FINDLEY S D, CANNON S, VARALA K, DU J C, MA J X, HUDSON M E, BIRCHLER J, STACEY G. A fluorescence in situ hybridization system for karyotyping soybean. Genetics, 2010, 185(3): 727-744. doi: 10.1534/genetics.109.113753
[21] DANILOVA T V, FRIEBE B, GILL B S. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma, 2012, 121(6): 597-611. doi: 10.1007/s00412-012-0384-7
[22] SUN J Y, ZHANG Z H, ZONG X, HUANG S W, LI Z Y, HAN Y H. A high-resolution cucumber cytogenetic map integrated with the genome assembly. BMC Genomics, 2013, 14(1): 461-468. doi: 10.1186/1471-2164-14-461
[23] YU F, WANG H Q, ZHAO Y Y, LIU R J, DOU Q W, DONG J L, WANG T. Karyotypic evolution of the Medicago complex: Sativa-caerulea-falcata inferred from comparative cytogenetic analysis. BMC Evolutionary Biology, 2017, 17(1): 104-120. doi: 10.1186/s12862-017-0951-x
[24] 马晓岗, 蒋礼玲, 徐媛君, 任建东. 青海高原地区作物种质资源的收集保护和创新利用进展. 植物遗传资源学报, 2015, 16(6): 1272-1276. MA X G, JIANG L L, XU Y J, REN J D. Progress of conversation and inovation of crop germplasm resources in Qinghai Plateau regions. Journal of Plant Genetic Resources, 2015, 16(6): 1272-1276.
[25] ANDRES R J, KURAPARTHY V. Development of an improved method of mitotic metaphase chromosome preparation compatible for fluorescence in situ hybridization in cotton. The Journal of Cotton Science, 2013, 17: 149-156.
[26] LIU R J, YU F, WEI L N, LIU B, LIU D M, DOU Q W. Development and application of transposable element-based chromosomal markers for the St Genome in Triticeae. Cytogenetic and Genome Research, 2019, 159(4): 215-224. doi: 10.1159/000504690
[27] CUADRADO A, JOUVE N. Novel simple sequence repeats (SSRs) detected by ND-FISH in heterochromatin of Drosophila melanogaster. BMC Genomics, 2011, 12: 205-219. doi: 10.1186/1471-2164-12-205
[28] DOU Q W, TANAKA H, NAKATA N, TSUJIMOTO H. Molecular cytogenetic analyses of hexaploid lines spontaneously appearing in octoploid Triticale. Theoretical and Applied Genetics, 2006, 114(1): 41-57. doi: 10.1007/s00122-006-0408-x
[29] LIU R J, WANG R R C, YU F, LU X W, DOU Q W. Characterization of genome in tetraploid StY Elymus (Triticeae: Poaceae) species using sequential FISH and GISH. Genome, 2017, 60(8): 679-685. doi: 10.1139/gen-2017-0046
[30] KUBIS S, SCHMIDT T, SEYMOUR H J. Repetitive DNA elements as a major component of plant genomes. Annals of Botany, 1998, 82: 45-55.
[31] YEN C, YANG J L, YANG Y. Hitoshi Kihara, Áskell Löve and the modern genetic concept of the genera in the tribe Triticeae (Poaceae). Acta Phytotaxonomica Sinica, 2005, 43: 82-93.
[32] ZENG DY, GUAN J T, LUO J T, HAO L B, LI Y Z, CHEN W S, ZHANG L Q, NING S Z, YUAN Z W, LI A L, ZHENG Y L, MAO L, LIU D C, HAO M. A transcriptomic view of the ability of nascent hexaploid wheat to tolerate aneuploidy. BMC Plant Biology, 2020, 20(1): 97-112. doi: 10.1186/s12870-020-2309-6
[33] ZHANG H K, BIAN Y, GUO X W, ZHU B, XU C M, QI B, LI N, RUSTGI S, ZHOU G, HAN F P, JIANG J M, WETTSTEIN S V, LIU B. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3447-3452.
[34] GUERRA M. Chromosome numbers in plant cytotaxonomy: Concepts and implications. Cytogenetic and Genome Research, 2008, 120(3/4): 339-350. doi: 10.1159/000121083
[35] 任建东, 王晓醒, 马晓岗, 马瑞强, 李长慧. 三江源7个糙毛以礼草植物学形态及遗传多样性分析. 草业科学, 2023, 40(1): 249-257. doi: 10.11829/j.issn.1001-0629.2022-0229 REN J D, WANG X X, MA X G, MA R Q, LI C H. Botanical morphology and genetic diversity analysis of seven Kengyilia hirsute germplasms in the areas of Sanjiangyuan. Pratacultural Science, 2023, 40(1): 249-257. doi: 10.11829/j.issn.1001-0629.2022-0229
-
图 1 顺序FISH和GISH探针在大颖草体细胞染色体上的荧光原位杂交结果
K1-a、K2-a、K3-a、K4-a和K5-a为第1轮S5 (红色)和(AAG)10 (绿色)探针的杂交信号。K1-b、K2-b、K3-b、K4-b和K5-b为第2轮糙缘拟鹅观草(红色)和沙芦草(绿色)基因组DNA的杂交信号。
Figure 1. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Kengyilia grandiglumis.
K1-a, K2-a, K3-a, K4-a, and K5-a: initially FISH probed with S5 (red) and (AAG)10 (green). K1-b, K2-b, K3-b, K4-b, and K5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Agropyron mongolicum (green).
图 2 顺序FISH和GISH探针在短芒披碱草体细胞染色体上的荧光原位杂交结果
E1-a、E2-a、E3-a、E4-a和E5-a为第1轮S5 (红色)和AAG (绿色)探针的杂交信号。E1-b、E2-b、E3-b、E4-b和E5-b为第2轮糙缘拟鹅观草(红色)和布顿大麦(绿色)基因组DNA的杂交信号。箭头和方框指示的是非整倍中的第43条染色体。
Figure 2. Physical mapping of sequential FISH and GISH probes on the mitotic chromosomes of Elymus breviaristatus.
E1-a, E2-a, E3-a, E4-a, and E5-a: initially FISH probed with S5 (red) and (AAG) 10 (green). E1-b, E2-b, E3-b, E4-b, and E5-b: sequentially GISH probed with genomic DNAs of Pseudoroegneria stipifolia (red) and Hordeum bogdanii (green). The arrow and box indicate chromosome 43 in aneuploid.
-
[1] 中国科学院中国植物志委员会. 中国植物志. 第九卷. 北京: 科学出版社, 1987: 6-116. Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora Reipublicae Popularis Sinicae. vol. 9. Beijing: Science Press, 1987: 6-116.
[2] 青海植物志编辑委员会, 青海植物志. 第四卷. 西宁: 青海人民出版社, 1996: 87-98. Editorial Committee of Flora of Qinghai. Flora of Qinghai. vol 4. Xining: Qinghai People’s House, 1996: 87-98.
[3] 唐俊伟, 乔安海, 马力, 贾顺斌, 王晓彤. 播种技术对大颖草产量及农艺性状的影响. 草地学报, 2019, 27(5): 1425-1430. TANG J W, QIAO A H, MA L, JIA S B, WANG X T. The effect of the rowing technologies on yield and agronomic characters of Roegeria grandigumis. Acta Agrestia Sinica, 2019, 27(5): 1425-1430.
[4] 施建军, 马玉寿, 董全民, 王彦龙, 王柳英. “黑土型”退化草地优良牧草筛选试验. 草地学报, 2007, 15(6): 543-549. SHI J J, MA Y S, DONG Q M, WANG Y L, WANG L Y. The selection experiment of fine forages in ‘Black Siol Type’ degraded grasslands. Acta Agrestia Sinica, 2007, 15(6): 543-549.
[5] 张利, 周永红, 郑有良, 张颖. 仲彬草属八个物种的核型. 植物分类与资源学报, 2005, 27(1): 81-86. ZHANG L, ZHOU Y H, ZHENG Y L, ZHANG Y. Karyotypes of 8 species in Kengyilia. Acta Botanica Yunnanica, 2005, 27(1): 81-86.
[6] DOU Q W, WANG R R-C, LEI Y T, YU F, LI Y, WANG H Q, CHEN Z G. Genome analysis of seven species of Kengyilia (Triticeae: Poaceae) with FISH and GISH. Genome, 2013, 56(11): 641-649. doi: 10.1139/gen-2013-0113
[7] ZENG J, ZHANG L, FAN X, ZHANG H Q, YANG R W, ZHOU Y H. Phylogenetic analysis of Kengyilia species based on nuclear ribosomal DNA internal transcribed spacer sequences. Biologia Plantarum, 2008, 52(2): 231-236. doi: 10.1007/s10535-008-0051-2
[8] LUO X M, TINKER N A, FAN X, ZHANG H Q, SHA L N, KANG H Y, DING C B, LIU J, ZHANG L, YANG R W, ZHOU Y H. Phylogeny and maternal donor of Kengyilia species (Poaceae: Triticeae) based on three cpDNA (matK, rbcL and trnH-psbA) sequences. Biochemical Systematics and Ecology, 2012, 44: 61-69.
[9] ZHANG L, ZHENG Y L, WEI Y M, LIU S G, ZHOU Y H. The genetic diversity and similarities among Kengyilia species based on random amplified microsatellite polymorphism (RAMP). Genetic Resources and Crop Evolution, 2005, 2(8): 1011-1017.
[10] WANG Q X, XIANG J S, GAO A N, YANG X M, LIU W H, LI X Q, LIU H L. Analysis of chromosomal structural polymorphisms in the St, P, and Y genomes of Triticeae (Poaceae). Genome, 2010, 3(3): 241-249.
[11] 任建东, 李凤珍, 徐媛君, 王晓醒, 马晓岗. 基于EST-SSR分子标记的青海高原以礼草属主要物种的遗传多样性分析. 植物遗传资源学报, 2016, 17(4): 663-670. REN J D, LI F Z, XU Y J, WANG X X, MA X G. Genetic diversity of the major varieties of Kengyilia in Qinghai Plateau based on EST-SSR markers. Journal of Plant Genetic Resources, 2016, 17(4): 663-670.
[12] 刘玉红. 我国11种披碱草的核型研究. 植物科学学报, 1985, 3(4): 325-330. LIU Y H. Studies on the karyotypes of 11 species of Elymus from China. Journal of Wuhan Botanical Research, 1985, 3(4): 325-330.
[13] YANG C R, BAUM B-R, CHEN W H, ZHANG H Q, LIU X Y, FAN X, SHA L N, KANG H Y, WANG Y, ZHOU Y H. Genomic constitution and taxonomy of the Chinese hexaploids Elymus cylindricus and E. breviaristatus (Poaceae: Triticeae). Botanical Journal of the Linnean Society, 2016, 182(3): 650-657. doi: 10.1111/boj.12469
[14] 顾晓燕, 郭志慧, 张新全, 周凯, 周朝杰, 符开欣, 刘新, 马啸. 短芒披碱草异位保护群体的表型多样性研究. 草业学报, 2015, 24(5): 141-152. doi: 10.11686/cyxb20150517 GU X Y, GUO Z H, ZHANG X Q, ZHOU K, ZHOU C J, FU K X, LIU X, MA X. Phenotypic variations in seven ex-situ conservation populations of Elymus breviaristatus. Acta Prataculturae Sinica, 2015, 24(5): 141-152. doi: 10.11686/cyxb20150517
[15] GU X Y, GUO Z H, MA X, BAI S Q, ZHANG X Q, ZHANG C B, CHEN S Y, PENG Y, YAN Y H, HUANG L K, ZHOU K, ZHOU C J, FU K X. Population genetic variability and structure of Elymus breviaristatus (Poaceae: Triticeae) endemic to Qinghai-Tibetan Plateau inferred from SSR markers. Biochemical Systematics and Ecology, 2015, 58: 247-256.
[16] LI J, ZHANG C B, CHEN S Y, JIANG K K, GUAN H, LIU W H. Characterization and application of EST-SSR markers developed from transcriptome sequences in Elymus breviaristatus (Poaceae: Triticeae). Genes, 2023, 14(2): 302. doi: 10.3390/genes14020302
[17] 顾晓燕, 郭智慧, 张新全, 刘新, 周朝杰, 马啸. 短芒披碱草SRAP-PCR体系的建立和优化. 中国农学通报, 2014, 30(30): 259-264. doi: 10.11924/j.issn.1000-6850.2014-1190 GU X Y, GUO Z H, ZHANG X Q, LIU X, ZHOU C J, MA X. Establishment and optimization of SRAP-PCR reaction system for Elymus breviaristatus. Chines Agricultural Science Bulletin, 2014, 30(30): 259-264. doi: 10.11924/j.issn.1000-6850.2014-1190
[18] BLANCATO J K. Fluorescence in Situ Hybridization. In The Principles of Clinical Cytogenetics. New Jersey: Humana Press, 1999: 443-471.
[19] KATO A, VEGA J M, HAN F P, LAMB J C, BIRCHLER J A. Advances in plant chromosome identification and cytogenetic techniques. Current Opinion in Plant Biology, 2005, 8(2): 148-154. doi: 10.1016/j.pbi.2005.01.014
[20] FINDLEY S D, CANNON S, VARALA K, DU J C, MA J X, HUDSON M E, BIRCHLER J, STACEY G. A fluorescence in situ hybridization system for karyotyping soybean. Genetics, 2010, 185(3): 727-744. doi: 10.1534/genetics.109.113753
[21] DANILOVA T V, FRIEBE B, GILL B S. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma, 2012, 121(6): 597-611. doi: 10.1007/s00412-012-0384-7
[22] SUN J Y, ZHANG Z H, ZONG X, HUANG S W, LI Z Y, HAN Y H. A high-resolution cucumber cytogenetic map integrated with the genome assembly. BMC Genomics, 2013, 14(1): 461-468. doi: 10.1186/1471-2164-14-461
[23] YU F, WANG H Q, ZHAO Y Y, LIU R J, DOU Q W, DONG J L, WANG T. Karyotypic evolution of the Medicago complex: Sativa-caerulea-falcata inferred from comparative cytogenetic analysis. BMC Evolutionary Biology, 2017, 17(1): 104-120. doi: 10.1186/s12862-017-0951-x
[24] 马晓岗, 蒋礼玲, 徐媛君, 任建东. 青海高原地区作物种质资源的收集保护和创新利用进展. 植物遗传资源学报, 2015, 16(6): 1272-1276. MA X G, JIANG L L, XU Y J, REN J D. Progress of conversation and inovation of crop germplasm resources in Qinghai Plateau regions. Journal of Plant Genetic Resources, 2015, 16(6): 1272-1276.
[25] ANDRES R J, KURAPARTHY V. Development of an improved method of mitotic metaphase chromosome preparation compatible for fluorescence in situ hybridization in cotton. The Journal of Cotton Science, 2013, 17: 149-156.
[26] LIU R J, YU F, WEI L N, LIU B, LIU D M, DOU Q W. Development and application of transposable element-based chromosomal markers for the St Genome in Triticeae. Cytogenetic and Genome Research, 2019, 159(4): 215-224. doi: 10.1159/000504690
[27] CUADRADO A, JOUVE N. Novel simple sequence repeats (SSRs) detected by ND-FISH in heterochromatin of Drosophila melanogaster. BMC Genomics, 2011, 12: 205-219. doi: 10.1186/1471-2164-12-205
[28] DOU Q W, TANAKA H, NAKATA N, TSUJIMOTO H. Molecular cytogenetic analyses of hexaploid lines spontaneously appearing in octoploid Triticale. Theoretical and Applied Genetics, 2006, 114(1): 41-57. doi: 10.1007/s00122-006-0408-x
[29] LIU R J, WANG R R C, YU F, LU X W, DOU Q W. Characterization of genome in tetraploid StY Elymus (Triticeae: Poaceae) species using sequential FISH and GISH. Genome, 2017, 60(8): 679-685. doi: 10.1139/gen-2017-0046
[30] KUBIS S, SCHMIDT T, SEYMOUR H J. Repetitive DNA elements as a major component of plant genomes. Annals of Botany, 1998, 82: 45-55.
[31] YEN C, YANG J L, YANG Y. Hitoshi Kihara, Áskell Löve and the modern genetic concept of the genera in the tribe Triticeae (Poaceae). Acta Phytotaxonomica Sinica, 2005, 43: 82-93.
[32] ZENG DY, GUAN J T, LUO J T, HAO L B, LI Y Z, CHEN W S, ZHANG L Q, NING S Z, YUAN Z W, LI A L, ZHENG Y L, MAO L, LIU D C, HAO M. A transcriptomic view of the ability of nascent hexaploid wheat to tolerate aneuploidy. BMC Plant Biology, 2020, 20(1): 97-112. doi: 10.1186/s12870-020-2309-6
[33] ZHANG H K, BIAN Y, GUO X W, ZHU B, XU C M, QI B, LI N, RUSTGI S, ZHOU G, HAN F P, JIANG J M, WETTSTEIN S V, LIU B. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3447-3452.
[34] GUERRA M. Chromosome numbers in plant cytotaxonomy: Concepts and implications. Cytogenetic and Genome Research, 2008, 120(3/4): 339-350. doi: 10.1159/000121083
[35] 任建东, 王晓醒, 马晓岗, 马瑞强, 李长慧. 三江源7个糙毛以礼草植物学形态及遗传多样性分析. 草业科学, 2023, 40(1): 249-257. doi: 10.11829/j.issn.1001-0629.2022-0229 REN J D, WANG X X, MA X G, MA R Q, LI C H. Botanical morphology and genetic diversity analysis of seven Kengyilia hirsute germplasms in the areas of Sanjiangyuan. Pratacultural Science, 2023, 40(1): 249-257. doi: 10.11829/j.issn.1001-0629.2022-0229



 下载:
下载: